By the Gigantum founding team (detailed below).
Today, we present Gigantum, an open source platform for creating and collaborating on computational and analytic work, complete with:
- Automated, high-resolution versioning of code, data and environment for reproducibility and rollback
- Work and version history illustrated in a browsable activity feed
- Streamlined environment management with customization via Docker snippets
- One-click transfer between laptop and cloud for easy sharing
- Seamless integration with development environments such as JupyterLab

We created Gigantum because modern science needs modern tools. Like most disciplines, the life sciences are now inextricably linked with the digital world of data and computation. As producers and consumers of knowledge, many researchers must employ new skills and practices in their daily work, with their activities ranging in complexity from computing summary statistics and model fitting, to developing sophisticated simulations and machine learning.
Concomitant with their power, digital analyses are accompanied by a wealth of frustrations. A host of seemingly non-deterministic factors can make the sharing of data and code challenging at best, and impossible at worst. The complexity of installing and maintaining customized tools and the difficulty of understanding ad hoc organizational conventions often impedes transparency and validation. The application of best practices, such as versioning and documentation of code, is a perennial chore. Even if one knows what current best practices are, employing them can be so time consuming and laborious that they are often ignored.
As scientists ourselves, we and our team struggled with having the time and skill to manage the code, data and environments necessary to be productive and incorporate the latest tools. So we designed and created a platform that reduces friction in our daily workflow, saves us time and stress, and captures valuable information that would typically be lost. It is specifically tailored to the type of “data science” that is increasingly a part of all sciences, not just the life sciences. We call the platform Gigantum.
We know there are already a variety of ways to address individual aspects of the overall challenge. Git, when combined with services like GitHub or GitLab, offers a structured way to work together and manage code. Tools such as Anaconda and Docker give varying degrees of environment management, replication and code portability, while platforms like Jupyter and RStudio provide interactive programming environments for exploration and development. There are also great tools like Binder and Stencila that help to provide reproducibility through the provision of computational execution contexts for digital lab notebooks and computationally rich scientific papers alike.
However, orchestrating and maintaining such a broad set of technologies requires skill and time, which end users may not have, and we saw no ergonomic way to manage enough of them at once. So we started developing Gigantum with the following broad goals:
- Significantly mitigate the labor and difficulties of installing and using tools locally
- Enable best practices for transparency, portability and reproducibility
- Allow the easy transfer of work between laptop and cloud, individual and institution
Additionally, there were some governing principles that we wanted the end product to follow:
- Automation is best for tasks that humans are too busy or lazy to do
- It should be easy to use, but still allow for highly customized packages and tools
- Work should be reproducible and portable when it is first created, not post hoc
- Capturing and communicating work products should include the who, what and when
- Meet people where they work, be it locally or in the cloud, and get out of their way
So, what is Gigantum?
The Gigantum Client is a local, browser-based application built on Docker that streamlines the tasks needed to make your work more intelligible, reproducible and portable. It allows you to quickly set up a customized development environment, work in JupyterLab and then share your work for publication or collaboration with just a few clicks. Automated, fine-grained versioning provides high-resolution rollback, and the entire history of the Project is accessible and illustrated in a detailed Activity Feed that includes a who/what/when timeline.
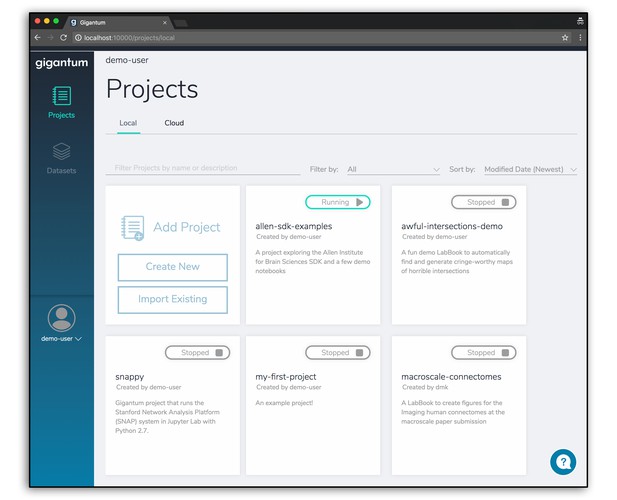
The Gigantum Client is a browser-based app that runs locally to manage the annoying and skill-intensive tasks needed to make your work versioned, documented, portable and reproducible. It automates and simplifies environment creation and high-resolution versioning in the context of an easy-to-use interface that lets you create and share work for storage, publication and easy collaboration. Right now, it integrates with JupyterLab as a development environment and we plan to integrate with tools such as RStudio and Stencila soon.
The Gigantum Client is open source (see our GitHub organization) and can be used locally by visiting gigantum.com/download to install. An online demo is available here and the following video shows how to get started.
Other than create portable, reproducible and documented Jupyter notebooks easily, what else can you do with Gigantum? At the moment, there are a few things you might use it for:
- Sharing your work with another Gigantum user for easy collaboration
- Complying with availability and reproducibility requirements of publishers and funders
- Getting access to interesting packages and tools from a variety of scientific projects
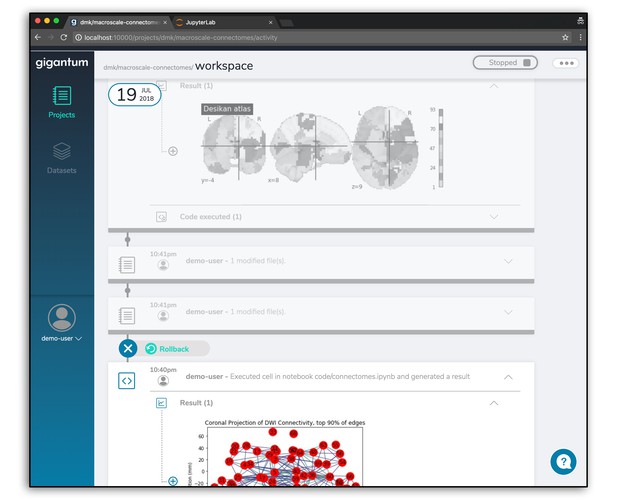
The high resolution and automated versioning of Gigantum makes rollback pretty simple. Any of the dots in the activity feed represent a particular configuration of data, code and environment that can be revisited by rolling back. You can learn more about rollback here.
Collaborating through Gigantum is pretty easy. You just publish your work to the cloud and add collaborators by username. They can download it, work on it and then sync it back. The versioning and rollback that are exposed through the Activity Feed make reproducibility an afterthought. They let anybody easily recreate the conditions (code/data/environment) that produced a given result, even if it is from many versions ago.
Finally, accessing interesting projects or complicated libraries that you may not know how to install yourself is easy with Gigantum. On our examples page, you can easily access the Allen Institute for Brain Science SDK to explore their data, as well as other custom tools for processing and analyzing brain imaging data. Or, you can use more general machine learning and analysis tools for things like manifold learning for millions of points or network analysis.
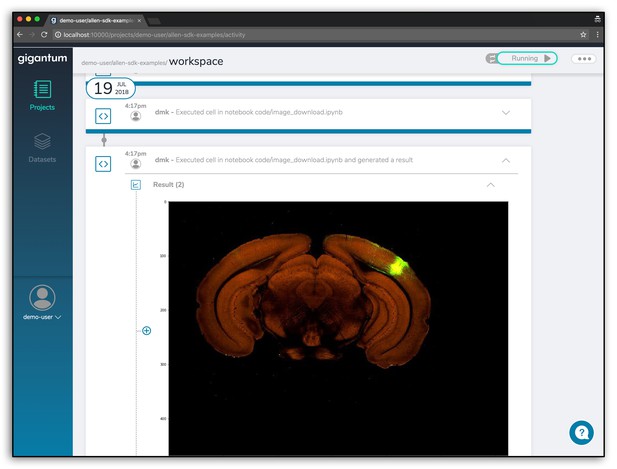
What is going on under the hood?
This is all possible because Gigantum bundles and versions code, data and environment into something we call a Gigantum Project, which practically is just an augmented Git repository. Projects are an open format for organizing and recording everything needed to be done and to then reproduce your work. They can be manually inspected using a file browser or Git, synced to the cloud repository, or exported and shared like ordinary files.
Gigantum creates new versions as you work, automatically making commits with additional data such as figure thumbnails and code snippets. Everything is visually rendered in the Project’s Activity Feed, so you won’t go blind looking through commit hashes and manually entered messages.
Another core component is automated Docker management and Jupyter integration. Gigantum creates a Dockerfile based on the user’s configuration choices, builds the container and bind mounts the desired content from the host machine for you. The Client automatically connects to Jupyter kernels and extracts information and creates versions without needing any user action.
A lot more went into creating the software. If you are curious about the details, see this blog post on Medium.
What is next?
At the moment, Gigantum lets you create, share and review work done in Jupyter notebooks in a reproducible and portable fashion. You can work locally but still share and collaborate using the Gigantum Cloud repository. Furthermore, features like the Activity Feed and rollback make sure you never forget where you were or lose important work. While this is already a useful set of features, we’ve got quite a few other things coming over the next year.
The most immediate update is that soon Gigantum Cloud will not just provide storage, but will also expose a searchable interface to view and find Projects. Users will be able to post their work for sharing or publication, letting anyone replicate their work with just a few clicks. Both public and private Projects will be supported, along with standard permission controls for individuals and eventually teams.
In the short term, we will also be adding new features to the local Client. For example, soon you will be able to search and filter the Activity Feed to quickly locate items. We’re also adding more development environments, including RStudio and Stencila. Additionally, the Client’s file browser capabilities are being expanded to include file previews. Finally, you will soon be able to link external Git repositories so you can easily use shared libraries managed by other platforms such as GitHub.
What is our basic model, as a company?
We develop software for scientists and data scientists because openness and reproducibility are important to us. The bottom line is that we want to provide a way for anybody, anywhere in the world, to create and share their work easily with collaborators and the rest of the world. There should be no barriers to accessing and using the work. That is why the Gigantum Client is MIT licensed and easy to use locally. We are committed to maintaining the open source Gigantum Client and releasing new innovations as we develop them.
We also know that any effective solution must utilize cloud resources. The cloud is how you reach people and access scale. That is why we are focused on making the transfer between laptop and cloud as seamless as possible. It’s also why the coming version of Gigantum Cloud will provide an open and outward-facing repository of public Projects for inspection and forking. We want it to be easy for everybody in the world to find exciting and useful work.
As (former) working researchers, we are acutely aware of the cost constraints that scientists and other individuals are subject to. In general, people can’t afford to burn budgets on cloud resources or large-scale enterprise software just because they are easier to use. That is why we are committed to providing a level of free services for the storing and sharing of Projects. But we want to stay sustainable and make it easy for anybody to access the full scale of cloud resources when they need to, and that is why we will also offer a paid tier for those that need real scale on occasion.
That is also why we want to make it as easy as possible for you to work locally. There is absolutely no point in paying for things when you don’t have to. Our ultimate goal is to simultaneously maintain a sustainable company, promote the democratization of science, and give on-demand access to the full power of the cloud. We think it is possible.
Conclusion
We created Gigantum to solve our own problems, but we hope that it helps to make data science and research easier, more reproducible and portable for everybody. We are looking for users to test it, play with it and provide feedback on both the current set of features and also desired capabilities that are missing.
If you use Gigantum (either the demo or the local install), please let us know! We want to hear what you like and don’t like, and we definitely want to help if you encounter any problems or bugs. Use the discussion forum to let us know what is on your mind and to see what other users have to say.
Gigantum Founders: Randal Burns, Dean Kleissas, Jacob Vogelstein, Joshua Vogelstein, Tyler Whitehouse
##
We welcome comments, questions and feedback. Please annotate publicly on the article or contact us at innovation [at] elifesciences [dot] org.
Do you know someone with an idea to improve the way research is shared and evaluated? They could build it with project funding, mentorship and exposure through the eLife Innovation Initiative. Get in touch.




