By Daniel Nüst, Institute for Geoinformatics at the University of Münster, Germany
One of the 13 projects at the eLife Innovation Sprint 2018 was an integration of Binder and Stencila. This article reports on the project's inception, building blocks, achievements at the sprint weekend and work conducted in the following months. Today, Binder has first-class Stencila support. You can open Stencila documents from any online code repository on mybinder.org with the click of a single button:
The idea and the sprint team
The eLife Innovation Sprint started with brief introductions by all participants. Some of them prepared pitches for project ideas, which quickly got little group discussions going. One table at the sprint attracted a few people with an interest in containerisation technology for research applications. Many ideas were floated and a helpful exchange around existing solutions and tools took place. When it was time to find a concrete task, two of the sprinters identified a worthwhile technological problem as their challenge for the next 1.5 days and the project "Jupyter+DAR compatibility exploration" started. Min from the Simula Research Laboratory, Norway, is a core developer of Binder and related tools. He was interested in getting to know the Stencila project and exploring the possibilities of having alternative user interfaces on Jupyter Hub. I myself, Daniel Nüst, from the o2r project at the Institute for Geoinformatics, Germany, work on reproducible computations in the geosciences and have a keen interest in learning more about the Binder platform. We were joined remotely by Nokome, the initiator and one of the developers of Stencila.
The building blocks
Stencila is an office suite for reproducible research documents. It provides an intuitive known word processor and spreadsheet user interface to scientists. Not all scientists are software developers ("coders"), or have the resources for learning all complex technologies which underpin today's software setups, nor should they be. Instead, they apply established software to answer important research questions, and Stencila makes reproducible research more accessible to them. Stencila is built upon Texture, a tool set for scientific content, which uses the Dar file format. Dar is designed for reproducible research publications, serving both the workflows of researchers using computational methods for data, and publishers using digital workflows for publication and preservation of scholarly journals.
A Binder makes it simple to generate reproducible computing environments from online repositories. The online service mybinder.org is the most prominent example of a platform based on the Binder project, a part of Project Jupyter. A user can run a Jupyter Notebook and other environments for their research projects, which are published in online repositories (such as GitHub or GitLab; see binder examples). In the spirit of the Unix philosophy, Binder combines several open-source tools to achieve this goal: repo2docker, for generating Dockerfiles and building Docker images from a software projects, JupyterHub for executing a Docker image and user-facing web portal in a cloud environment, and BinderHub for gluing the above together.
A Dockerfile is a human and machine-readable recipe for setting up a computational environment, which is just a fancy way of saying "installing and configuring software". Dockerfiles are used by the popular Docker container software. They can be built into an executable image, which is portable between host computers. These properties make containers very interesting for capturing and sharing research involving data and software.
While containers have become a commodity for developers, researchers still struggle to grasp and control the complexity of computational environments. This is where the two building blocks join: running Stencila as part of a Binder helps researchers to communicate their work openly, to collaborate effectively with other scientists and to ensure the high quality and transparency of their workflow and findings.
The challenge
As Min and I formulated our goals in the sprint project form, the project was heavy with software titles:
"Compatibility between Jupyter Notebooks / Binder and DAR / Texture / Stencila
Our goal was "[..] to connect them so that users can edit reproducible documents (DAR files) as part of a Binder project" with the following objectives: (i) understanding DAR [Dar Format], (ii) launching Stencila Editor on Binder (potentially not launching anything else, i.e. without the Jupyter Notebook start page), and (iii) repo2docker support for DAR files. The project was also part of the Mozilla Global Sprint 2018; see mozilla/global-sprint#317.
The solution
It took more than just the 1.5 days in Cambridge to really fulfil this challenge. First we describe the crucial breakthroughs that were actually made at the sprint, then the updates that have happened between then and now.
Sprint breakthrough
Min and I started by taking a close look at an existing solution, namely the integration of RStudio based on nbrsessionproxy, i.e. the "Notebook R session proxy". We learned two things:
- a Jupyter notebook extension can be used to add a menu item to the Jupyter user interface (UI)
- a component is needed to route the traffic between the browser-based user interface and the server-side software
The first attempts utilised Binder's feature of manually defining a bespoke Dockerfile (see a first attempt) and later also a postBuild script to install and configure all software. It was my first task to transfer the first finding for Stencila. After setting up a local development environment and learning Jupyter/Binder, it just needed small adjustments to selected files from nbrsessionproxy to achieve this (see commit from the second day):
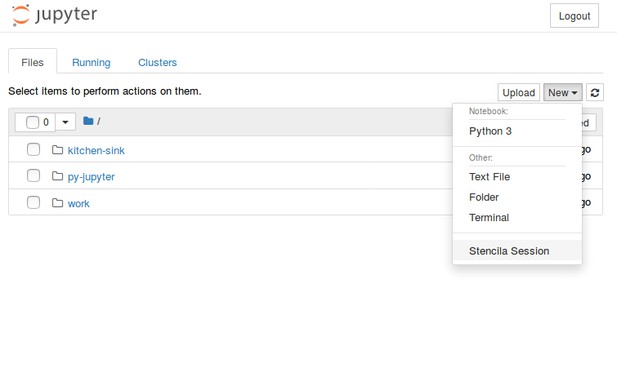
Min took on the second task while at the same time figuring out which parts of Stencila we really needed, and how to glue them together. He wrote a hard-wired proxy using Python and added some JavaScript/HTML files to serve Dar files and the Stencila UI itself.
Connecting Stencila to Jupyter kernels
Stencila has "execution contexts" (the equivalent of Jupyter's "kernels") for R, Python, SQL, Javascript (in the browser), and Node.js. Execution contexts differ from kernels in a number of ways, including local execution and dependency analysis of cells. Both of these are necessary for the reactive, functional execution model of Stencila Articles and Sheets.
We could install these execution contexts in the Docker image. However, Stencila also has a JupyterContext which acts as a bridge between Stencila's application programming interface and Jupyter kernels. So, since the base jupyter/minimal-notebook image already has a Jupyter kernel for Python installed in it, we decided to use that. This did mean however, that some of the reactive aspects of the Stencila UI wouldn't work as expected.
We included the stencila-node Node.js package in the Docker image, which provides the JupyterContext as well as a NodeContext (for executing Javascript) and a SqliteContext (for executing SQL).
We first used Stencila's development build to run the JavaScript app using node make -w -s -d /our/own/dir, but struggled slightly to configure the file storage, i.e. the dar-server, to use the directory we wanted, and to run it in a full path configured by us instead of make.js starting the dar-server relative to __dirname. Eventually we ended up implementing our own minimal JavaScript module (i.e. an npm package) that ran (i) the dar-server and (ii) a static file server for the app using the distribution files (i.e. the dist directory). This gave us control of the paths and let us get rid of complex development features, such as substance-bundler.
We also made our own version of app.js, removing the virtual file storage (vfs; used to seamlessly integrate examples) and instead defaulting to a file system (fs) storage, because that is what is needed for Jupyter. In the same line, we built our own index.html (based on example.html) to serve as the entry page. This allowed us to directly render a single Dar document, instead of a listing of examples, and to use our own app.js. Relevant path configurations comprised the local storage path as well as the URLs used by the client, accessing the dar-server through the nbserverproxy.
At the end of the first day, the wiring was all there so we could open a repository, and the Stencila document was shown! But the interactive execution of code cells did not work yet :-/.
Thanks to an international time-zone-difference-powered "overnight" contribution, Min and I got a big surprise on Friday morning: Nokome added the Stencila Node.js host for Jupyter execution context support, so that Python cells could be executed by connecting to the Jupyter Kernel, which of course was already there. In doing so, he returned the "surprise" he had when learning about the project. The added "host" provides the single gateway for code-cell contents to be forwarded to the respective execution contexts. Nokome showed that everything worked with the obligatory screenshot:
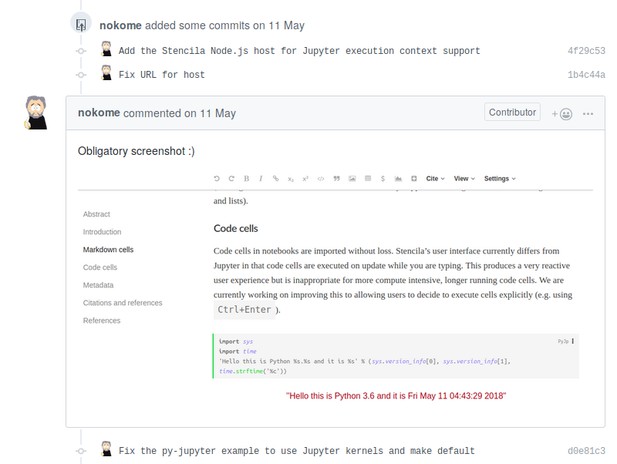
Since you can run any commit in a Binder, you can also try out that particular state from the repository yourself:
The second day of the sprint involved many iterations of improvements, including changes to repo2docker. These updates could not simply be thrown upon mybinder.org, so Min set up a test server for the demonstrations on the sprint's final day. I continued my work on supporting R code cells, albeit with small contributions to the Stencila codebase that meant I could not complete this task in time.
The sprint ended with presentations by all projects, some of which are still going strong today, for example Appstract, Citation Gecko and PREreview. The results were truly awesome, ranging from ambitious concepts, case studies, design concepts and completely new tools with great user experience designs, to technical demonstrators. It's an easy guess where on the spectrum our project can be placed...You're invited to catch a glimpse of the sprint, its results and the people behind all of it on Twitter under the hashtag #eLifeSprint and read the project roundup.
The following screencast and Binder link show the status at the end of the sprint: a Stencila document could be opened on a bespoke Binder deployment and the contained Python code could be interactively edited. The code is rerun on the server and the figure updated.
You can view the Python example document by appending ?archive=py-jupyter to the URL of Stencila in the Binder, e.g. https://hub.mybinder.org/.../stencila/?archive=py-jupyter.
Consolidation
A couple of weeks after the sprint, a second, less-intensive development period started. I continued my work on adding support for the R context, and also managed to get plain Python cells running (see pull requests #15 and #16). Min restructured the whole project and gave it the name it still bears: nbstencilaproxy – a Jupyter notebook server extension and proxy for Stencila.
The project’s GitHub repository holds a Python module with the Jupyter notebook server and "non-server" extensions of the same name, and a bundled JavaScript module, also of the same name.
The Python module allows proper versioned installation, dependency management and installation from an established software repository. It takes care of the plumbing between the user interface and the services in the background, so that the Binder is viewable over one port in the browser, while the many different background components run on their own ports. The "non-server" extension adds the "Stencila session" menu entry and conveniently lives in the same directory structure as the server extension.
The JavaScript module manages the required JavaScript dependencies and provides a well-defined structure for the code files. It serves the Dar document and provides access to the Stencila host (see above).
While complex at first sight, this modularity hopefully makes maintenance for future developments and new collaborators easier. For now, the JavaScript module and its installation are bundled with the Python module instead of being published independently, because the code and configuration are very much specific to the Jupyter integration.
Min also extended repo2docker with automatic detection of Dar documents (as part of a "build pack"), so that no configuration is required for the most common use cases. As with most Binder repositories, a user could simply open a Dar document on Binder and trust the required environment to provide all required software.
On July 20, the nbstencilaproxy was published on PyPI and, on August 1, the new developments made it into repo2docker. Soon after, Stencila was available for all users on mybinder.org, which was a great achievement for a little project started at a community sprint. However, the big announcement was still not made, since some things were still hard-wired and, to use R for example, the author of a repository had to manually add a configuration file, although the information that R needs is already part of the Stencila document.
The last mile
In October, I took on the final tasks of writing this blog post and fixing the R installation, including the automatic detection of the required execution contexts of a given Dar document. This included some housekeeping in nbstencilaproxy and, more importantly, new contributions to repo2docker (thanks to Tim for his review and help) to (i) properly detect the languages used in a Stencila document, (ii) extend the R build pack to install R if it is used in a Stencila document, and (iii) add documentation and tests. repo2docker now detects Dar documents based on their manifest.xml files and uses the location of the first discovered one as the base directory to start Stencila. If a Dar manifest is found, then nbstencilaproxy is installed and the languages are extracted from code cells from the document. Authors can install extra dependencies using the repo2docker's existing mechanisms.
I also created a few example repositories to provide a starting point for users. The Binder team generously welcomed the changes to mybinder.org and the examples to the binder examples organisation on GitHub. The following repositories contain single or multiple Stencila documents with code chunks in different programming languages.
https://github.com/binder-examples/stencila-py contains Python code cells, using both the Jupyter and plain Python execution contexts:
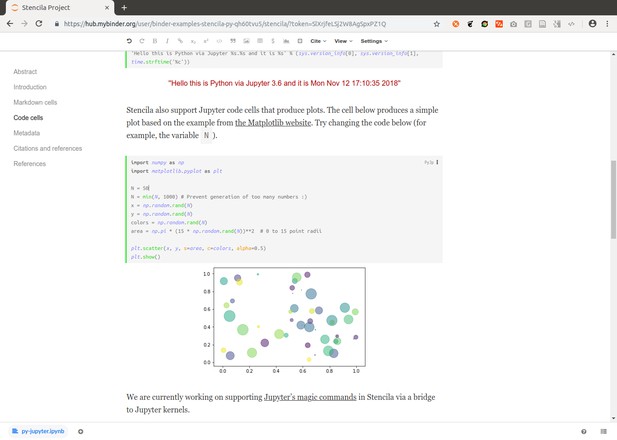
https://github.com/binder-examples/stencila-r contains R code cells and two plots:
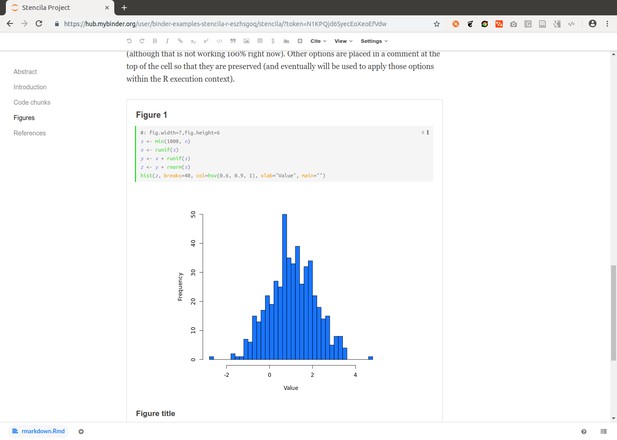
https://github.com/binder-examples/stencila-multi demonstrates how to access specific Dar projects if multiples are found within a repository.
In each case, you can see the available execution environments by clicking on the icon in the bottom-right corner.
One of the cool features of Stencila are the reactive cells, as demonstrated in a tweet following the feature release:
"Thanks to @nordholmen working on @stencila support for https://t.co/Zlj6FrYgBw you now have reactive cells with Python code on @mybinderteam! Give it a go https://t.co/ToIuQPq0Fy pic.twitter.com/Wjyf1kiH9B"
— Tim Head (@betatim) November 12, 2018
Summary and outlook
Thanks for reading so far! This blog post is a long-planned write up of the history of the tool and decisions mostly relevant to developers, but it’s also a demonstration of the power that the open-source and open-science community can foster. Many people are working together on the (technological) challenges of science today towards full research transparency and reproducibility, even if we use computers to an unprecedented level. Many small contributions on "side projects" such as these can make a difference, and connecting these two great projects will hopefully help to solve some problem in science down the road.
What's next? While there are no concrete plans, there are some ideas listed on the project's issue tracker, such as automatic conversion from a Jupyter notebook to Dar when there is no Dar archive in a repository. In any case, you can keep an eye out on GitHub for projects being tagged as Stencila and Binder, and join the public Stencila and Binder chats to stay in touch or get help. We look forward to seeing scientists use nbstencilaproxy for communicating their work and the new challenges that come with it.
This post was originally published here on Github.
We welcome comments, questions and feedback. Please annotate publicly on the article or contact us at innovation [at] elifesciences [dot] org.
Do you have an idea or innovation to share? Send a short outline for a Labs blogpost to innovation [at] elifesciences [dot] org.
For the latest in innovation, eLife Labs and new open-source tools, sign up for our technology and innovation newsletter. You can also follow @eLifeInnovation on Twitter.




