Peer review process
Not revised: This Reviewed Preprint includes the authors’ original preprint (without revision), an eLife assessment, public reviews, and a provisional response from the authors.
Read more about eLife’s peer review process.Editors
- Reviewing EditorJoshua CorbinChildren's National Hospital, Washington, United States of America
- Senior EditorJonathan CooperFred Hutch Cancer Center, Seattle, United States of America
Reviewer #1 (Public Review):
Summary:
SUFU modulates Sonic hedgehog (SHH) signaling and is frequently mutated in the B-subtype of SHH-driven medulloblastoma. The B-subtype occurs mostly in infants, is often metastatic, and lacks specific treatment. Yabut et al. found that Fgf5 was highly expressed in the B-subtype of SHH-driven medulloblastoma by examining a published microarray expression dataset. They then investigated how Fgf5 functions in the cerebellum of mice that have embryonic Sufu loss of function. This loss was induced using the hGFAP-cre transgene, which is expressed in multiple cell types in the developing cerebellum, including granule neuron precursors (GNPs) derived from the rhombic lip. By measuring the area of Pax6+ cells in the external granule cell layer (EGL) of Sufu-cKO mice at postnatal day 0, they find Pax6+ cells occupy a larger area in the posterior lobe adjacent to the secondary fissure, which is poorly defined. They show that Fgf5 RNA and phosphoErk1/2 immunostaining are also higher in the same disrupted region. Some of the phosphoErk1/2+ cells are proliferative in the Sufu-cKO. Western blot analysis of Gli proteins that modulate SHH signaling found reduced expression and absence of Gli1 activity in the region of cerebellar dysgenesis in Sufu-cKO mice. This suggests the GNP expansion in this region is independent of SHH signaling. Amazingly, intraventricular injection of the FGFR1-2 antagonist AZD4547 from P0-4 and examined histologically at P7 found the treatment restored cytoarchitecture in the cerebella of Sufu-cKO mice. This is further supported by NeuN immunostaining in the internal granule cell layer, which labels mature, non-diving neurons, and KI67 immunostaining, indicating dividing cells, and primarily found in the EGL. The mice were treated beginning at a timepoint when cerebellar cytoarchitecture was shown to be disrupted and it is indistinguishable from control following treatment. Figure 3 presents the most convincing and exciting data in this manuscript.
Sufu-cKO do not readily develop cerebellar tumors. The authors detected phosphorylated H2AX immunostaining, which labels double-strand breaks, in some cells in the EGL in regions of cerebellar dysgenesis in the Sufu-cKO, as was cleaved Caspase 3, a marker of apoptosis. P53, downstream of the double-strand break pathway, the protein was reduced in Sufu-cKO cerebellum. Genetically removing p53 from the Sufu-cKO cerebellum resulted in cerebellar tumors in 2-month old mice. The Sufu;p53-dKO cerebella at P0 lacked clear foliation, and the secondary fissure, even more so than the Sufu-cKO. Fgf5 RNA and signaling (pERK1/2) were also expressed ectopically.
The conclusions of the paper are largely supported by the data, but some data analysis need to be clarified and extended.
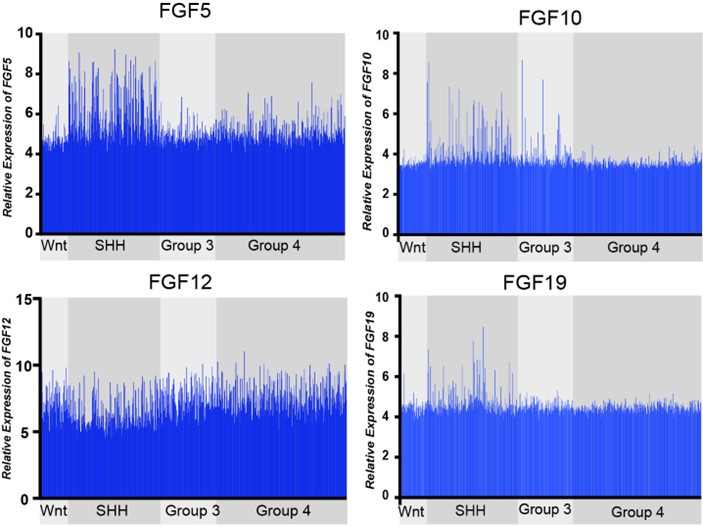
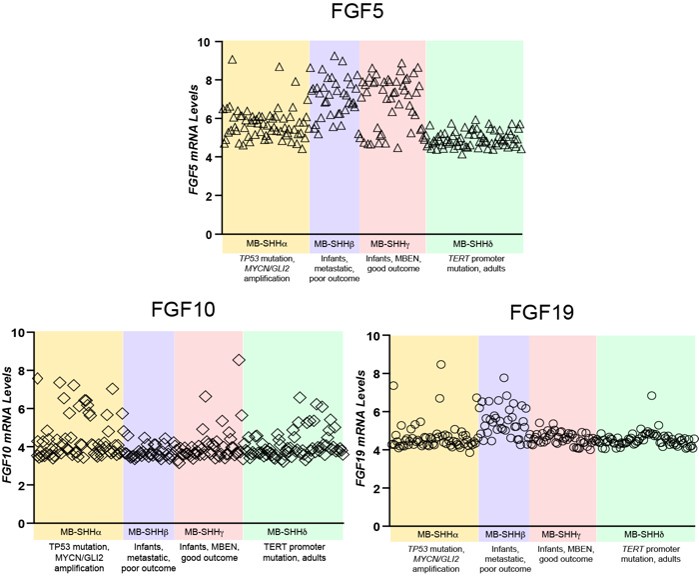
(1) The rationale for examining Fgf5 in medulloblastoma is not sufficiently convincing. The authors previously reported that Fgf15 was upregulated in neocortical progenitors of mice with conditional loss of Sufu (PMID: 32737167). In Figure 1, the authors report FGF5 expression is higher in SHH-type medulloblastoma, especially the beta and gamma subtypes mostly found in infants. These data were derived from a genome-wide dataset and are shown without correction for multiple testing, including other Fgfs. Showing the expression of other Fgfs with FDR correction would better substantiate their choice or moving this figure to later in the manuscript as support for their mouse investigations would be more convincing.
(2) The Sufu-cKO cerebellum lacks a clear anchor point at the secondary fissure and foliation is disrupted in the central and posterior lobes. It would be helpful for the authors to review Sudarov & Joyner (PMID: 18053187) for nomenclature specific to the developing cerebellum.
(3) The metrics used to quantify cerebellar perimeter and immunostaining are not sufficiently described. It is unclear whether the individual points in the bar graph represent a single section from independent mice, or multiple sections from the same mice. For example, in Figures 2B-D. This also applies to Figure 3C-D.
(4) The data on Fgf5 RNA expression presented in Figure 2E are not sufficiently convincing. The perimeter and cytoarchitecture of the cerebellum are difficult to see and the higher magnification shown in 2F should be indicated in 2E.
(5) The data presented in Figure 3 are not sufficiently convincing. The number of cells double positive for pErk and KI67 (Figure 3B) are difficult to see and appear to be few, suggesting the quantification may be unreliable.
(6) The data presented in Figure 4F-J would be more convincing with quantification. The Sufu;p53-dKO appears to have a thickened EGL across the entire vermis perimeter, and very little foliation, relative to control and single cKO cerebella. This is a more widespread effect than the more localized foliation disruption in the Sufu-cKO.
(7) Figure 5 does not convincingly summarize the results. Blue and purple cells in sagittal cartoon are not defined. Which cells express Fgf5 (or other Fgfs) has not been determined. The yellow cells are not defined in relation to the initial cartoon on the left.
Reviewer #2 (Public Review):
Summary:
Mutations in SUFU are implicated in SHH medulloblastoma (MB). SUFU modulates Shh signaling in a context-dependent manner, making its role in MB pathology complex and not fully understood. This study reports that elevated FGF5 levels are associated with a specific subtype of SHH MB, particularly in pediatric cases. The authors demonstrate that Sufu deletion in a mouse model leads to abnormal proliferation of granule cell precursors (GCPs) at the secondary fissure (region B), correlating with increased Fgf5 expression. Notably, pharmacological inhibition of FGFR restores normal cerebellar development in Sufu mutant mice.
Strengths:
The identification of increased FGF5 in subsets of MB is novel and a key strength of the paper.
Weaknesses:
The study appears incomplete despite the potential significance of these findings. The current paper does not fully establish the causal relationship between Fgf5 and abnormal cerebellar development, nor does it clarify its connection to SUFU-related MB. Some conclusions seem overstated, and the central question of whether FGFR inhibition can prevent tumor formation remains untested.
Reviewer #3 (Public Review):
Summary:
The interaction between FGF signaling and SHH-mediated GNP expansion in MB, particularly in the context of Sufu LoF, has just begun to be understood. The manuscript by Yabut et al. establishes a connection between ectopic FGF5 expression and GNP over-expansion in a late-stage embryonic Sufu LoF model. The data provided links region-specific interaction between aberrant FGF5 signaling with the SHH subtype of medulloblastoma. New data from Yabut et al. suggest that ectopic FGF5 expression correlates with GNP expansion near the secondary fissure in Sufu LoF cerebella. Furthermore, pharmacological blockade of FGF signaling inhibits GNP proliferation. Interestingly, the data indicate that the timing of conditional Sufu deletion (E13.5 using the hGFAP-Cre line) results in different outcomes compared to later deletion (using Math1-cre line, Jiwani et al., 2020). This study provides significant insights into the molecular mechanisms driving GNP expansion in SHH subgroup MB, particularly in the context of Sufu LoF. It highlights the potential of targeting FGF5 signaling as a therapeutic strategy. Additionally, the research offers a model for better understanding MB subtypes and developing targeted treatments.
Strengths:
One notable strength of this study is the extraction and analysis of ectopic FGF5 expression from a subset of MB patient tumor samples. This translational aspect of the study enhances its relevance to human disease. By correlating findings from mouse models with patient data, the authors strengthen the validity of their conclusions and highlight the potential clinical implications of targeting FGF5 in MB therapy.
The data convincingly show that FGFR signaling activation drives GNP proliferation in Sufu, conditional knockout models. This finding is supported by robust experimental evidence, including pharmacological blockade of FGF signaling, which effectively inhibits GNP proliferation. The clear demonstration of a functional link between FGFR signaling and GNP expansion underscores the potential of FGFR as a therapeutic target in SHH subgroup medulloblastoma.
Previous studies have demonstrated the inhibitory effect of FGF2 on tumor cell proliferation in certain MB types, such as the ptc mutant (Fogarty et al., 2006)(Yaguchi et al., 2009). Findings in this manuscript provide additional support suggesting multiple roles for FGF signaling in cerebellar patterning and development.
Weaknesses:
In the GEO dataset analysis, where FGF5 expression is extracted, the reporting of the P-value lacks detail on the statistical methods used, such as whether an ANOVA or t-test was employed. Providing comprehensive statistical methodologies is crucial for assessing the rigor and reproducibility of the results. The absence of this information raises concerns about the robustness of the statistical analysis.
Another concern is related to the controls used in the study. Cre recombinase induces double-strand DNA breaks within the loxP sites, and the control mice did not carry the Cre transgene (as stated in the Method section), while Sufu-cKO mice did. This discrepancy necessitates an additional control group to evaluate the effects of Cre-induced double-strand breaks on phosphorylated H2AX-DSB signaling. Including this control would strengthen the validity of the findings by ensuring that observed effects are not artifacts of Cre recombinase activity.
Although the use of the hGFAP-Cre line allows genetic access to the late embryonic stage, this also targets multiple celltypes, including both GNPs and cerebellar glial cells. However, the authors focus primarily on GNPs without fully addressing the potential contributions of neuron-glial interaction. This oversight could limit the understanding of the broader cellular context in which FGF signaling influences tumor development.