Peer review process
Not revised: This Reviewed Preprint includes the authors’ original preprint (without revision), an eLife assessment, public reviews, and a provisional response from the authors.
Read more about eLife’s peer review process.Editors
- Reviewing EditorAlex NechiporukOregon Health and Science University, Portland, United States of America
- Senior EditorDidier StainierMax Planck Institute for Heart and Lung Research, Bad Nauheim, Germany
Reviewer #1 (Public review):
Summary:
Palardy and colleagues examine how transcription factors of the SoxB1 family alter patterning within the zebrafish posterior lateral line primordium and subsequent formation of neuromast organs along the body of the developing fish. They describe how expression of soxb genes changes when Wnt and Fgf signaling pathways are altered, and in addition, how outputs of these signalling pathways change when soxb gene expression is disrupted. Together, experiments suggest a model where the expression of SoxB genes counteracts Wnt signaling. Support comes from the combined inhibition of both pathways, partially restoring the pattern of neuromast deposition. Together, the work reveals an additional layer of control over Wnt and Fgf signals that together ensure proper posterior lateral line development.
Strengths:
The authors provide a clear analysis of changes in RNA expression after systematic manipulation of gene expression and signaling pathways to construct a plausible model of how Sox factors regulate primordium patterning.
Weaknesses:
There is little attempt to capture the variation of expression patterns with each manipulation. Photomicrographs are examples, with little quantification.
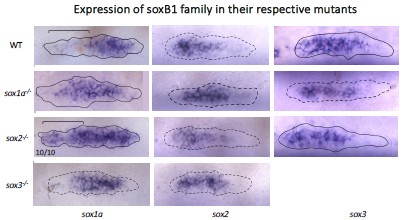
While the combined loss of soxb functions shows more severe phenotypes, it is not exactly clear what underlies the apparent redundancy. It would be helpful if the soxb gene family member expression was reported after loss of each. Expression of sox1a is shown in sox2 mutants in Figure 4, but other combinations are not reported. This additional analysis would clarify whether there are alterations in expression that influence apparent redundancy.
Reviewer #2 (Public review):
Summary:
This manuscript seeks to determine the molecular basis of tissue patterning in the collectively migrating cells of the zebrafish posterior lateral line primordium. In particular, the authors examine the cross-regulation of canonical Wnt signaling, Fgf signaling, and the SoxB1 family members Sox1a, Sox2, and Sox3 in the migrating primordium. Using a combination of mutant lines, morphino (MO) knock down, pharmacological inhibition, and dominant-negative inhibition, the authors propose a model in which Sox2 and Sox3 in the trailing region of the primordium restricts Wnt signaling to the leading region, facilitating the formation of rosettes and the deposition of the first formed neuromast downstream of Fgf pathway activity. In contrast, sox1a is expressed in the leading region of the primordium, and the sox1ay590 -/- mutant shows little phenotype on its own. Together, the authors propose a multistep signaling loop that regulates tissue patterning during lateral line collective cell migration.
Strengths:
The zebrafish posterior lateral line primordium is a well-established model for the study of collective cell migration that is useful for genetic manipulation and live imaging. The manuscript seeks to understand the complex reciprocal regulation of signaling pathways that regulate tissue patterning of collectively migrating cells.
Weaknesses:
(1) The primary tools used in this study are inadequate to support the author's conclusions.
A. The authors state that the phenotype of the sox2y589 homozygous mutant line described in this manuscript changed across generations, but do not specify which generation is used for any given experiment. The sox2y589 mutant line is not properly verified in this manuscript, which could be done by examining ant-Sox2 antibody labeling, Western blot analysis, or complementation to the existing sox2x50 line described in Gou et al., 2018a and Gou et al., 2018b. There are also published sox1a mutant lines Lekk, et al., 2019.
B. The authors acknowledge that the sox2 MO1 used in this manuscript also alters sox3 function, but do not redo the experiments with a specific sox2 MO. In addition, the authors show that the anti-Sox2 and anti-Sox3 antibody labeling is reduced but not absent in sox2 MO1 and sox3 MO-injected embryos, but do not show antibody labeling of the sox2 MO and sox3 MO-double injected embryos to determine if there is an additional knockdown.
C. The authors examine RNA in situ hybridization patterns of sox2 and sox3 following various manipulations, but do not use anti-Sox2 and anti-Sox3 antibody labeling, which would provide more quantifiable information about changes in patterning.
(2) The manuscript lacks important experimental details and appropriate quantification of results.
A. It is unclear for most of the experiments described in this manuscript how many individual embryos were examined for each experiment and how robust the results are for each condition. Only Figure 3 includes information about the numbers for each experiment, and in all cases, the experimental manipulations are not fully penetrant, and there is no statistical analysis.
B. It is not clear at what stage most of the RNA in situ hybridizations were performed.
C. The manuscript lacks quantification of many of the experiments, making it difficult to conclude their significance.
Reviewer #3 (Public review):
Summary:
This study aims to understand the molecular underpinnings of the complex process of periodic deposition of the neuromast organs of the embryonic posterior lateral line (PLL) sensory system in zebrafish. It was previously established that Fgf signaling in the trailing zone of the migrating PLL primordium is key to protoneuromast establishment, while Wnt signaling in the leading zone must be downregulated to allow new Fgf signaling-dependent protoneuromasts to form. Here, the authors evaluate the role of three SoxB transcription factors (Sox1a, Sox2, and Sox3) in this complex process, generating two novel CRISPR mutants as part of their study. They interrogate the interplay of the SoxB genes with the Fgf and Wnt signaling pathways during PLL primordium migration, using a combination of genetics, knockdown, and imaging approaches, including live time-lapse studies. They report a key role for the SoxB genes in regulating the pace of protoneuromast maturation as the primordium migrates, thus ensuring appropriate deposition and spacing of the neuromast organs.
Strengths:
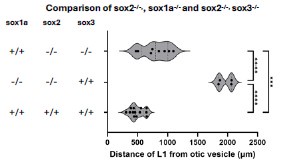
Strengths of the study are the careful quantitative analysis. based on imaging approaches, of the impact of mutation or knockdown of SoxB genes, coupled with the use of heat shock inducible dominant negative strategies to address how SoxB genes interact with Wnt and Fgf signaling. Functional analyses convincingly uncover a SoxB regulatory network that serves to limit Wnt activity, as directly read out with a live Wnt reporter. The finding that Wnt inhibition (achieved using pharmacological reagents) rescues the SoxB deficiency phenotype provides compelling evidence of the centrality of the Wnt pathway in mediating SoxB function. Use of atoh1 markers to track the stages of development of the neuromasts provides an effective approach to following their maturation, and allows the authors to explore how SoxB/Wnt interplay ultimately translates into the establishment of functional neuromasts. Finally, loss of Sox2 function, together with loss of either Sox1a or Sox3, blocks maturation of the neuromasts, clearly establishing redundancy between these SoxB family genes.
The concepts introduced and explored in this study - of complex gene networks that work within a dynamic cellular environment to enable self-organization and ultimately stabilization of cell fate choices-provide a useful conceptual framework for future studies. This study is therefore of relevance to understanding the morphogenesis of self-organizing tissues more broadly.
Weaknesses:
A minor weakness is the use of SoxB morpholino (MO) knockdown reagents, which are interspersed with mutant analyses. Although the stable mutants are available, they would be challenging to couple with the reporter transgenes used for some of the experiments, providing a reasonable rationale for the use of MO reagents (although the authors don't overtly provide this rationale). Moreover, reduced penetrance of the Sox2 mutants over multiple generations is noted, but no detailed explanation for this finding is offered.
Given that the expression patterns of Sox1a and Sox3 are not merely different but are largely reciprocal, the mechanistic basis of their very similar double mutant phenotypes with Sox2 remains opaque. Related to this, the authors discuss that Sox1a/Sox2 double knockdown produces a more severe phenotype than Sox2/Sox3 double knockdown, yet this difference is not obviously reflected in the data, some of which is not shown.