Peer review process
Not revised: This Reviewed Preprint includes the authors’ original preprint (without revision), an eLife assessment, public reviews, and a provisional response from the authors.
Read more about eLife’s peer review process.Editors
- Reviewing EditorMartin DenzelAltos Labs, Cambridge, United Kingdom
- Senior EditorDavid RonUniversity of Cambridge, Cambridge, United Kingdom
Reviewer #1 (Public Review):
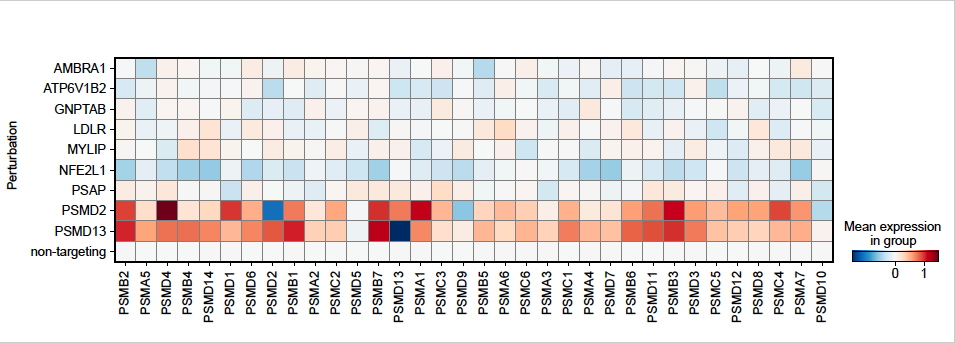
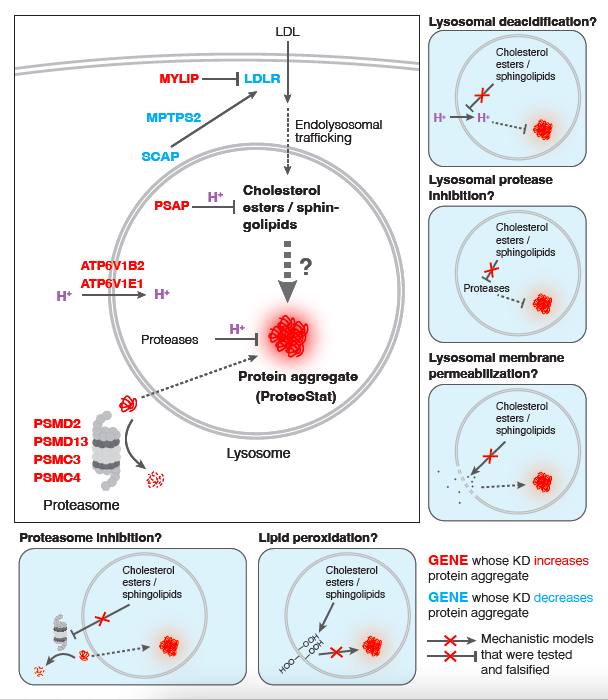
In this manuscript, Yong and colleagues link perturbations in lysosomal lipid metabolism with the generation of protein aggregates resulting from proteosome inhibition. The main tool used is the ProteoStat stain to assess protein aggregate burden in native cells (i.e. cells under no exogenous or endogenous stress). They initially use CRISPR-based genome-wide screens to identify several genes that affect this aggregate burden. Interestingly, knockdown of genes involved in lysosomal acidification was a major signature which led to identification of other culprit lysosome-associated genes that included ones involved in lipid metabolism. Subsequent CRISPR screen focused on lipidomic analysis led to identification of sphingolipid and cholesterol esters as lipid classes with effects on proteostasis. Despite using various tools of lysosomal function, acidity, permeability, etc, the authors couldn't identify the link between lysosomal lipid metabolism and protein aggregate formation. Nevertheless, the interrelationship of these two processes was the overall conclusion of this manuscript.
Although this work is interesting and thought-provoking, their approach to identify novel pathways involved in proteostasis is limited and this weakens the contribution of the paper in its current form.
Reviewer #2 (Public Review):
It is certainly an interesting observation that lipid homeostasis influences proteostasis, although this need not be considered so surprising given that many fundamental cellular processes are interconnected. The paper is deserves to be read, but the level of general interest would be greatly enhanced if the authors were able to take the story further mechanistically. This might be too much of an ask, but they should go further in excluding one very attractive alternative model: effects on proteasome activity. This explanation should be addressed definitively because the transcription factor that regulates proteasome subunit gene expression (Nrf1/NFE2L1) is processed in the ER and is therefore well placed to be influenced by membrane conditions, and because it is shown here that proteasome inhibition increase ProteoStat puncta. Indeed, some years ago it was published that Nrf1/NFE2L1 is inhibited within the ER membrane by cholesterol, and a more recent paper showed that in C. elegans it is activated by oleic acid through effects on ER membrane homeostasis and lipid droplet formation. The authors address proteasome activity only by using a dye that is not referenced. Here a much more solid answer is needed. In general, most conclusions in the paper rely essentially solely on ProteoStat assays. The entire study would be greatly strengthened if the authors incorporated biochemical or other modalities to substantiate their results.
The presentation would be improved greatly if the authors provided diagrams illustrating the pathways implicated in their results, as well as their models. As it is the paper falls flat at the end of the results in the absence of a mechanism to explain their findings. Diagrams would be helpful for focusing the reader on what IS learned from the work, which is important.