Peer review process
Revised: This Reviewed Preprint has been revised by the authors in response to the previous round of peer review; the eLife assessment and the public reviews have been updated where necessary by the editors and peer reviewers.
Read more about eLife’s peer review process.Editors
- Reviewing EditorYongliang YangShanghai University of Medicine and Health Sciences, Shanghai, China
- Senior EditorCaigang LiuShengjing Hospital of China Medical University, Shenyang, China
Reviewer #1 (Public Review):
In this study, Jiamin Lin et al. investigated the potential positive feedback loop between ZEB2 and ACSL4, which regulates lipid metabolism and breast cancer metastasis. They reported a correlation between high expression of ZEB2 and ACSL4 and poor survival of breast cancer patients, and showed that depletion of ZEB2 or ACSL4 significantly reduced lipid droplets abundance and cell migration in vitro. The authors also claimed that ZEB2 activated ACSL4 expression by directly binding to its promoter, while ACSL4 in turn stabilized ZEB2 by blocking its ubiquitination. While the topic is interesting, there are several concerns with the study:
1. My concern regarding the absence of appropriate thresholds or False Discovery Rate (FDR) adjustments for the RNA-seq analysis has not been addressed, leading to incorrect thresholds and erroneous identification of significant signals.
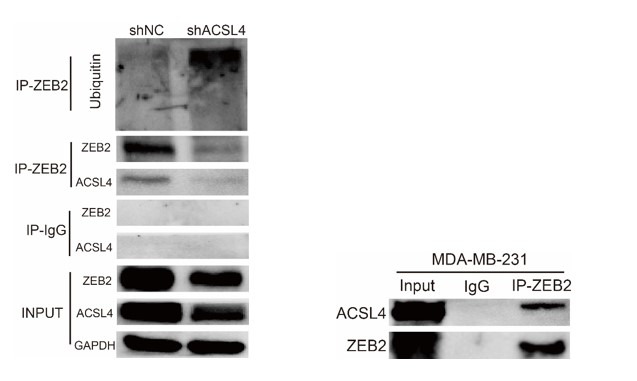
2. In Figure 3B and C, it appears that the knockdown efficiency of ACSL4 is inadequate in these cells, which contradicts the Western blot results presented in Figure 2F.
3. Regarding Figure 6, the discovery of consensus binding sequences (CACCT) for ZEB2 alone is insufficient evidence to support the direct binding of ZEB2 to the ACSL4 promoter.
4. For Figure 7E, there are multiple bands present, and it appears that ZEB2-HA has been cropped, which should ideally be presented with unaltered raw data. Please provide the uncropped raw data.
5. In Figure 7C, the author claimed to have used 293T cells for the ubiquitin assay, which are not breast cancer cells. Moreover, the efficiency of over-expression differs between ZEB2 and ACSL4 in 293T cell lines. Performing the experiment in an unrelated cell line to justify an important interaction is not acceptable.
Reviewer #2 (Public Review):
In this study, the authors validated a positive feedback loop between ZEB2 and ACSL4 in breast cancer, which regulates lipid metabolism to promote metastasis.
Overall, the study is original, well structured, and easy to read.
Reviewer #3 (Public Review):
The manuscript by Lin et al. reveals a novel positive regulatory loop between ZEB2 and ACSL4, which promotes lipid droplets storage to meet the energy needs of breast cancer metastasis.
Author Response
The following is the authors’ response to the original reviews.
On behalf of my co-authors, we thank you very much for giving us the opportunity to revise our manuscript entitled “A positive feedback loop between ZEB2 and ACSL4 regulates lipid metabolism to promote breast cancer metastasis” (manuscript number: eLife-RP-RA-2023-87510).
We would like to convey our appreciation to you and the expert reviewers for your valuable time and effort in reviewing and improving our work. We are grateful for the constructive comments raised by the six expert reviewers. We have studied the reviewer’s comments carefully and have accordingly conducted additional experiments as recommended. We have made the following revisions point by point. We found that our work was substantially strengthened by addressing these points.
Reviewer #1 (Public Review):
In this study, Jiamin Lin et al. investigated the potential positive feedback loop between ZEB2 and ACSL4, which regulates lipid metabolism and breast cancer metastasis. They reported a correlation between high expression of ZEB2 and ACSL4 and poor survival of breast cancer patients, and showed that depletion of ZEB2 or ACSL4 significantly reduced lipid droplets abundance and cell migration in vitro. The authors also claimed that ZEB2 activated ACSL4 expression by directly binding to its promoter, while ACSL4 in turn stabilized ZEB2 by blocking its ubiquitination. While the topic is interesting, there are several major concerns with the study and its conclusions are not convincing.
- Figure 1A, the clinical relevance or biological significance of drug-resistant luminal breast cancer cell lines with metastatic cancer is questionable. Additionally, the RNA-seq analysis lacked multiple test correction for differential gene expression analysis, and no fold-change cut-off was used, leading to incorrect thresholds and wrongly identified significant signals.
We appreciate the reviewer’s valuable questions to improve our manuscript. We identified many EMT related transcription factors such as ZEB2, SNAIL, TWIST, etc. was up-regulated in drug-resistant cells, so we hypothesized that drug-resistant cells may undergone EMT and acquire metastatic capability. The drug-resistant cells used in this study had already been proved and examined in the previous studies of our research team as follows:
(1) Zheng FM, Long ZJ, Hou ZJ et al., A novel small molecule aurora kinase inhibitor attenuates breast tumor-initiating cells and overcomes drug resistance. Mol Cancer Ther. 2014 Aug;13(8):1991-2003.
(2) Yang N, Wang C, Wang Z, et al., FOXM1 recruits nuclear Aurora kinase A to participate in a positive feedback loop essential for the self-renewal of breast cancer stem cells. Oncogene. 2017 Jun 15;36(24):3428-3440.
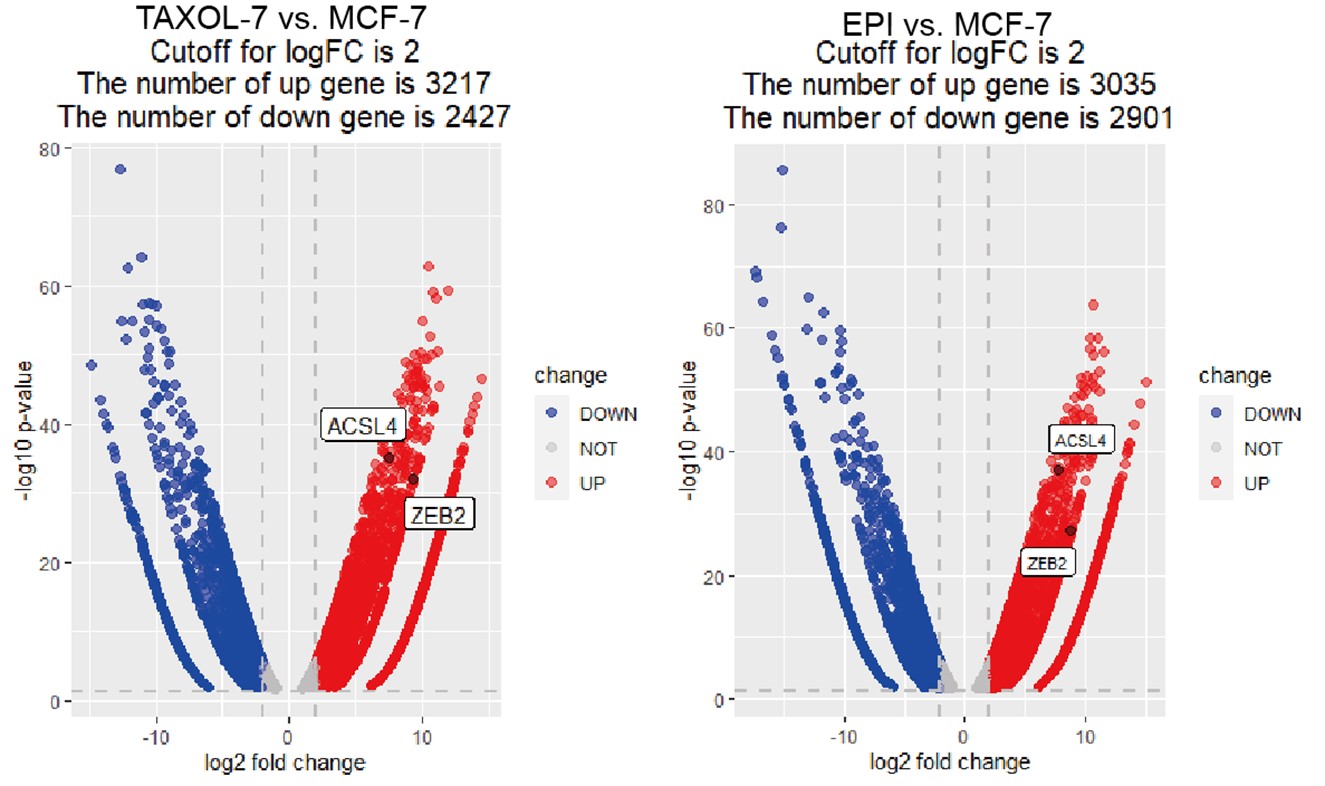
For the second question, we used the fold-change cut-off in RNA-seq analysis and the fold change was over 1.5-fold and the adjust P value is less than 0.05. To make it more clearly, we have reset the cut off with a |log2FC|2 and p<0.05 and generated the volcano Plot using R4.3.0 software for differentially expressed genes as follows in Author response image 1. The results showed 3217 and 3035 up-regulated genes in TAXOL-resistant and EPI-resistant cells respectively, along with 2427 (TAXOL) and 2901 (EPI) down-regulated genes. Both ACSL4 and ZEB2 were up-regulated in two cell lines. We have put the figure in the new supplementary Fig S2.
Author response image 1.
- Figure 1D-E, the clinical associations between ACSL4 and ZEB2 overexpression and poor patient survival are not justified. The authors used an old web tool, the Kaplan-Meier plotter database, based on microarray data, to perform the analysis. The reviewer repeated the analysis and found that multiple microarray probes for ZEB2 were available, leading to opposite results when different probes were selected. The reviewer also repeated the analysis using more reliable TCGA RNA-seq data and found no correlation between ASCL4 or ZEB2 expression and post-progression survival.
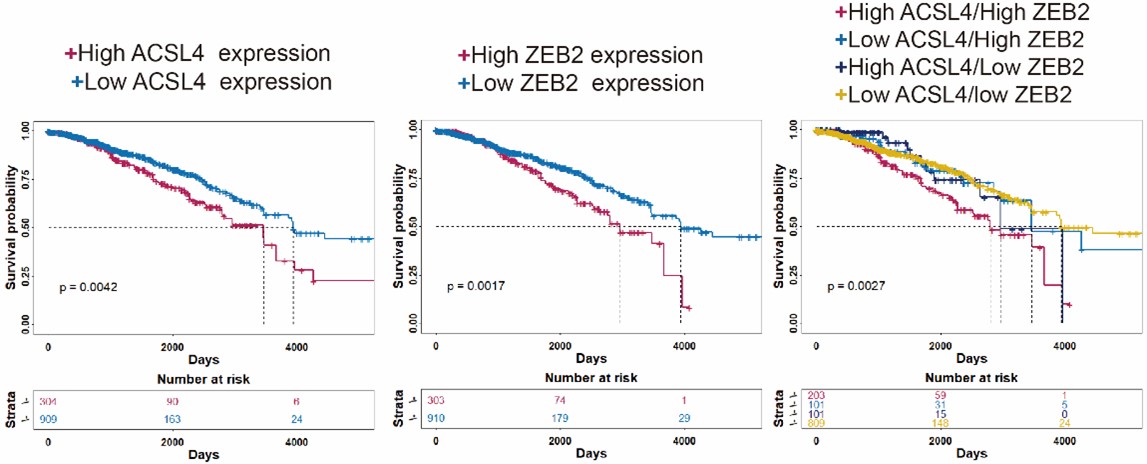
We appreciate the reviewer’s thoughtful questions. The Kaplan-Meier plotter database (http://kmplot.com/analysis/) we used is handled by a PostgreSQL server, which integrates gene expression and clinical data simultaneously including GEO, EGA and TCGA data. We used auto-select best cutoff for the the Kaplan-Meier analysis. Due to the web tool is old, we repeated the Kaplan-Meier survival analysis using R4.3.0 software and split the patients in TCGA database according to the third quartile expression (new Fig. 1D-F). The results also show that patients with high expression of ACSL4 and/or ZEB2 have relatively worse prognosis as follows in Author response image 2 (p<0.01):
Author response image 2.
- Figure 1I relied on IHC to support the negative correlation between ACSL4 and Erα expression, but the small sample size limits the power to establish the relationship and the results are not definitive without further replication or biological investigation. The authors should provide more detailed and comprehensive analysis, including appropriate statistical tests, to ensure the findings are robust and reliable.
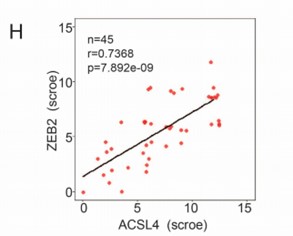
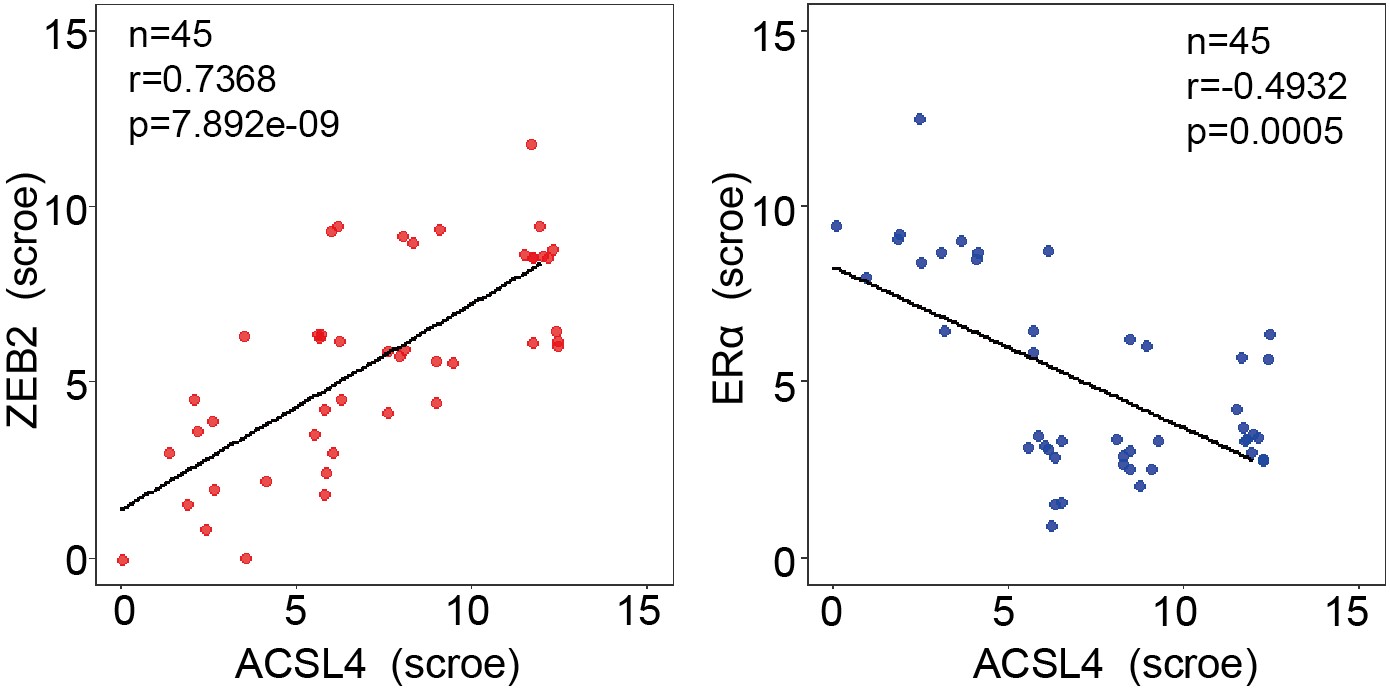
We appreciate the reviewer’s suggestion. To better understand the positive correlation between ACSL4 and ZEB2 expression, we add up to 45 breast cancer cases for IHC analysis and the correlation is shown as follows in Author response image 3 (new Fig. 1 H):
Author response image 3.
- Figure 3B-C lacks justification of the differences by showing only one field without any internal control for exposure. The reviewer suggests to show additional fields where cells with both efficiently and inefficiently knocked-down are present, to justify the robustness of the results. This can also be achieved by mixing control and knockdown cells.
We totally understand the reviewer's concern. Thank you for pointing out this problem. The lower magnification field of view is shown as follows and it includes both efficiently and inefficiently knocked-down cells. We have changed the Fig. 3B and C as follows in Author response image 4:
Author response image 4.
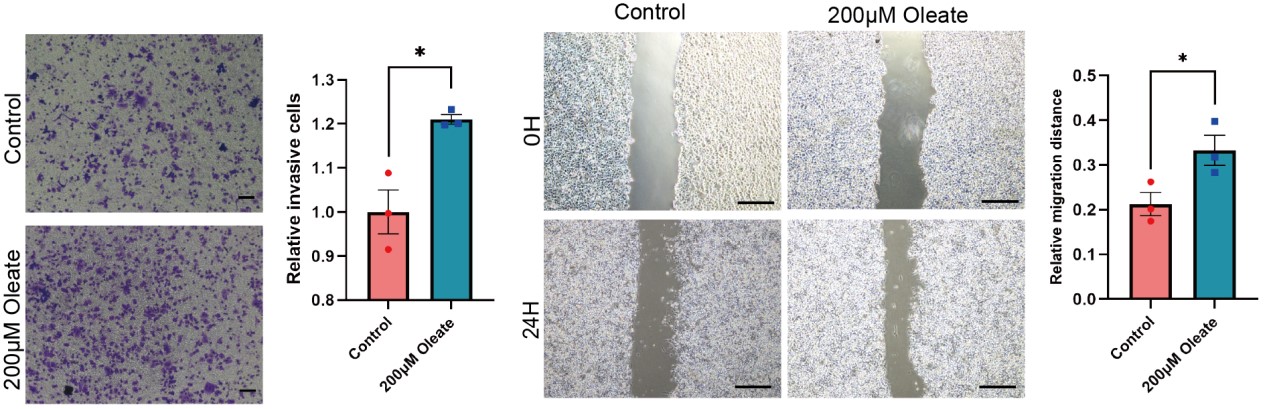
- Figure 4A-D, oleate-induced cell migration is a well-documented feature across different cancer types. To make it more relevant to the current study, the authors should examine multiple cell lines with high and low ZEB2/ACSL4 expression to determine the underlying relevance.
We appreciate the reviewer’s comments and performed the suggested experiments. To better determine the role of oleic acid and ACSL4 on cell migration, we use MCF-7 cell line, which has low ZEB2/ACSL4 expression, to test the influence of oleate on the cell migration. Transwell and Wound healing assays revealed that oleic acid treated MCF-7 cells also exhibited enhanced invasive and metastatic capacities compared with control cells. This results indicates that oleate induces cell migration in MCF-7 cells may via mechanisms other than ACSL4. We have added the results to the new Supplementary Fig. 8 as follows in Author response image 5.
Author response image 5.
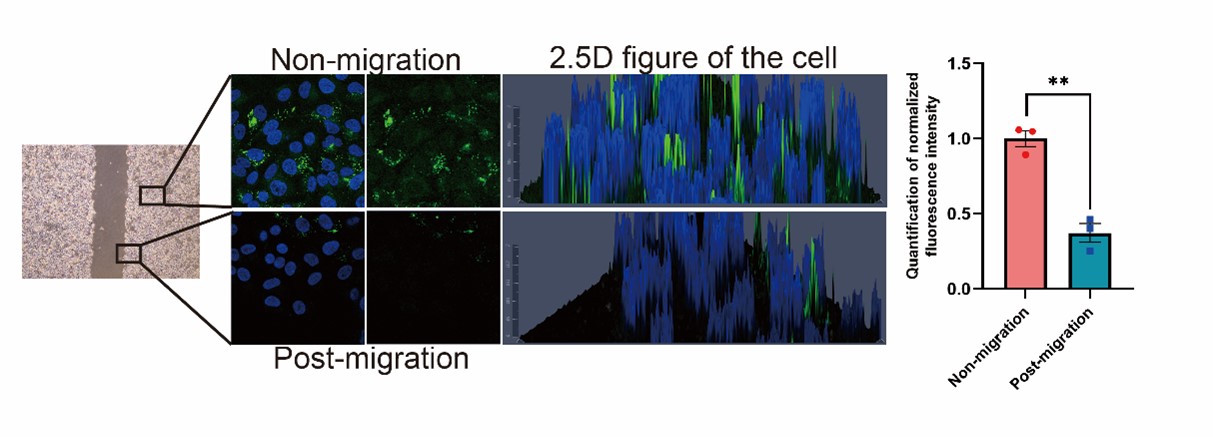
- Figure 4E, it is difficulty to conclude that cancer cells utilize stored lipids during migration to fuel metastasis based on current data. Do you see any evidence of lipid signal decreasing in the leading edge of the scratch wound-healing migration assay? The authors should also compare signals between unmigrated and migrated cells in the transwell assay.
We appreciate the reviewer’s constructive suggestion. We performed the wound-healing migration assay and observed that the lipid signal was obviously decreased in the leading edge of the scratch, as shown in the Author response image 6 (New Fig. 4E). In the transwell experiment, the cells which migrated to the lower side of the chamber after 24 hours showed decreased lipid signals (Fig. 4F). All these results indicates that lipid is utilized during migration.
Author response image 6.
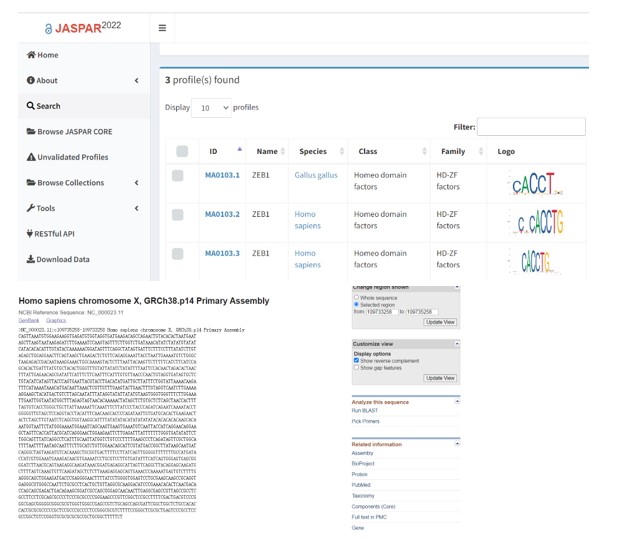
- Figure 6 warrants a genome-wide ChIP-seq to justify direct regulation of ASCL4 promoter by ZEB2. The reviewer’s analysis of publicly available ZEB2 ChIP-seq in multiple cell types detected no ZEB2 binding signaling within {plus minus} 5 kb of ASCL4 promoter.
We thank the reviewer for the concern. We found that the breast cancer cells are not included in some data base, such as Cistrome Data Browser, which is a resource of human and mouse cis-regulatory information derived from ChIP-seq, DNase-seq and ATAC-seq chromatin profiling assays. Due to that different cell type may have totally different mechanisms, that’s why the ZEB2 binding signaling cannot be found within ASCL4 promoter in some cells.
We searched JASPAR data base (https://jaspar.genereg.net/), which is an open-access database of non-redundant transcription factor (TF) binding profiles, and found the consensus binding sequences (CACCT) of ZEB (zinc finger E-box binding homeobox) transcription family were within the 2kb of ASCL4 promoter as follows in Author response image 7.
Author response image 7.
- Figure 7 presents a series of self-contradictory results. Figure 7C, why no significant change in ZEB2-MYC expression was observed in the presence of ACSL4 and/or HA-Ubi? In Figure 7 E&G, why robust ACSL4 expression is present in the control group in E but not in (G)? Additionally, why there is no degradation in ZEB2 baseline level over time in the shACSL4 group in E? These raise severe concerns about the data quality.
We appreciate the reviewer to point out these problems.
Response to question 1: In fig. 7C, we used 293T cell for the ubiquitin assay and it is not a breast cancer cells. The efficiency of over-expression is different between ZEB2 and ACSL4 in 293T cell lines.
Response to question 2: Because the expression of ACSL4 is low in MCF-7 and is high in MDA-MB-231 cells. In Figure 7E (New Fig. 7G), we used MDA-MB-231 cells for the control and ACSL4 knockdown cells. In Figure 7G (New Fig. 7I), we used MCF-7 cells for the control and ACSL4 over-expressed cells. We have also revised the figure legend of Fig.7 as follows:
I, The stability of ZEB2 protein was detected by CHX treatment assay in control or ACSL4 over-expressed MCF-7 cells. GAPDH was used as the internal loading control.
Response to question 3: Because knockdown of ACSL4 also significantly decreased the mRNA level of ZEB2 (New Fig. 7A), so the baseline levels of ZEB2 in the shACSL4 group (New Fig. 7G) were very low and degradation is not obvious.
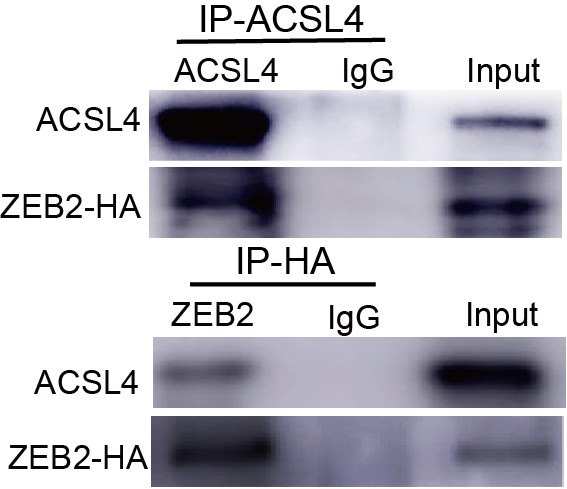
- Figure 7D, the IP result of ACSL4 is not justified as there is no enrichment of ACSL4 in the IP compared to input. With the current data, it is hard to justify that there is any direct interaction. Moreover, based on IF data in Figure 3B-C, ACSL4 is exclusively localized in the cytoplasm, while ZEB2 is exclusively localized in the nucleus. It is hard to believe there is any direct interaction and mutual regulation.
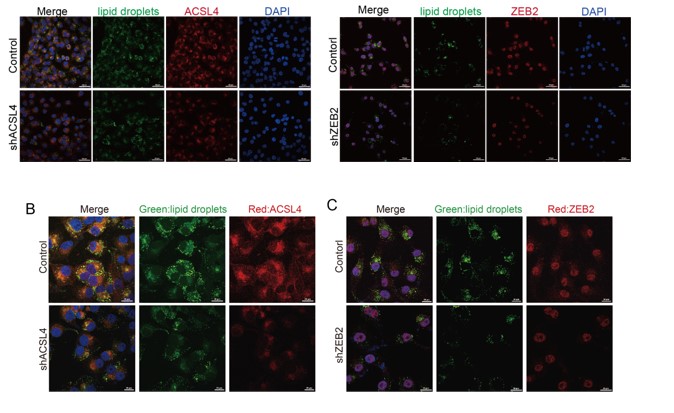
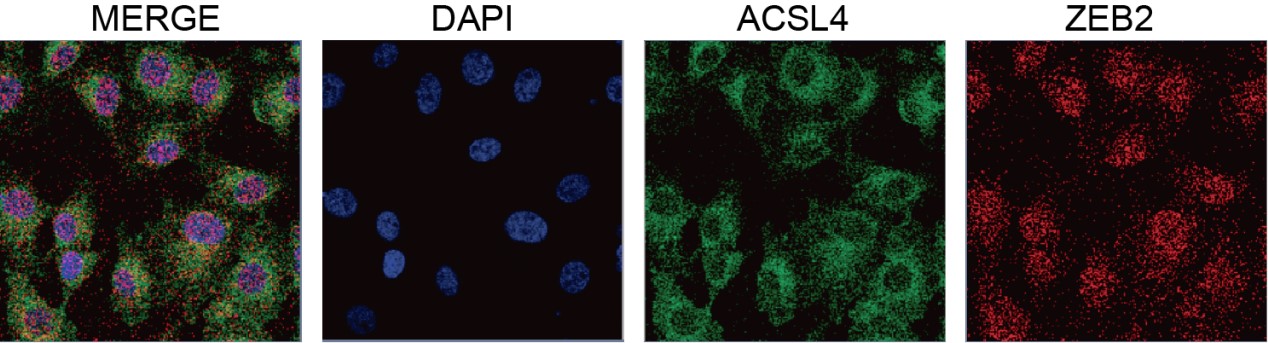
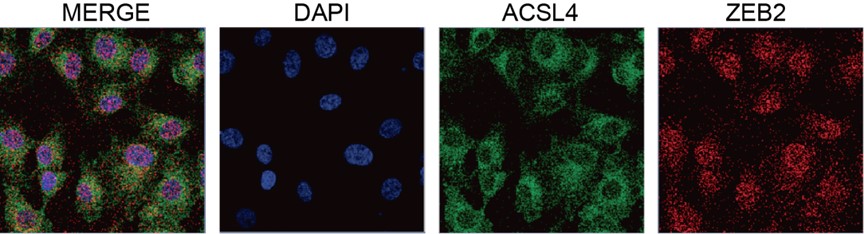
We appreciate the reviewer’s thoughtful questions. We have repeated the IP assay and found that the enrichment of ACSL4 was observed in the IP process and added to new Fig. 7E as follows in Author response image 8. We also repeated the immunofluorescence assay in the MDA-MB-231 cells. We observed that ZEB2 can also be found in the cytoplasm and co-localized with ACSL in some certain regions of the cytoplasm as follows in Author response image 9 (Supplementary Fig. S11):
Author response image 8.
Author response image 9.
Reviewer #2 (Public Review):
In this study, the authors validated a positive feedback loop between ZEB2 and ACSL4 in breast cancer, which regulates lipid metabolism to promote metastasis.
Overall, the study is original, well structured, and easy to read. Despite the reliability of the data discussed in this article, there are still some deficiencies that need to be addressed through further explanation.
Major issues:
- The authors demonstrated that ACSL4 regulates ZEB2 not only via a post-transcriptional mechanism but also via a transcriptional mechanism. The authors have not provided a comprehensive explanation of the specific mechanism in this paper. Therefore, it is recommended that the author delve into the potential mechanisms in the discussion section. For example, related mechanisms affecting ZEB2 ubiquitination degradation, as well as factors affecting ZEB2 upstream transcriptional regulation, etc.
We appreciate the positive comments and constructive suggestion from the reviewer. We have added the following paragraph in the second paragraph of the discussion section :
Interestingly, our RNA-seq data revealed that some ubiquitin E3 ligases, such as FBXO4, UBE3C, NEDD4, RBX1 etc. were significantly reduced in ACSL4 knockdown cells (Fig. S12). This result indicated that ACSL4 may reduce the ubiquitin of ZEB2 via down-regulating ubiquitin E3 ligase. Additionally, we found that ACSL4 promoted ZEB transcription as the mRNA level of ZEB2 was significantly reduced after ACSL4 knockdown. A recent study reported that LD-derived lipolysis provide acetyl-CoA for the epigenetic regulation of gene transcription. We observed that ACSL4 can also promote FAO, which generates acetyl-CoA for the epigenetic regulation. It is likely that ACSL4 regulates the ZEB2 mRNA level via lipid-epigenetic reprogramming mechanism, which is worth studying in the future.
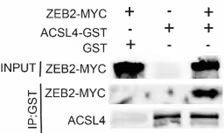
- To further clarify the interaction of ZEB2 and ACSL4, it is best to perform in vitro glutathione-S-transferase (GST) pulldown assay and immunofluorescence assay.
We appreciate the reviewer’s suggestion. We performed GST pull-down assay to examine whether ZEB2 and ACSL4 form a complex. GST pull-down assay confirmed the interaction of ZEB2 and ACSL4 as follows in Author response image 10 (Supplementary Fig. S10). We also performed immunofluorescence assay and found that ZEB2 was co-localized with ACSL in some certain regions of the cytoplasm as follows in Author response image 11. (Supplementary Fig. S11):
Author response image 10.
Author response image 11.
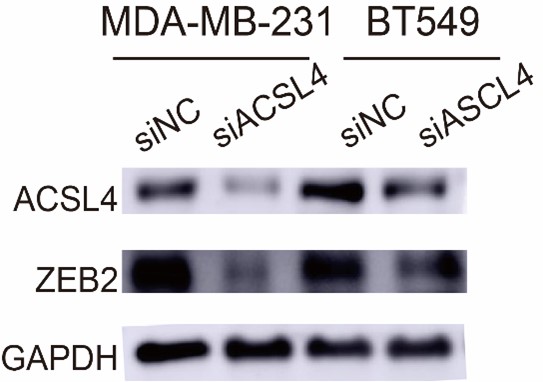
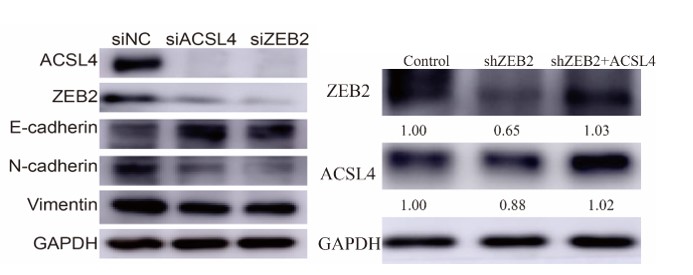
- In Figure 7B, the protein level of ZEB2 seems not to be altered in BT549 BCSC cell line after the depletion of ACSL4.
We appreciate the reviewer to point out this problem. The protein level of ZEB2 in BT549 BCSC cell is not abundant as MDA-MB-231. We repeated the experiment and found that ZEB2 was reduced after the depletion of ACSL4 in BT549. We have replaced the Fig.7B as follows in Author response image 12:
Author response image 12.
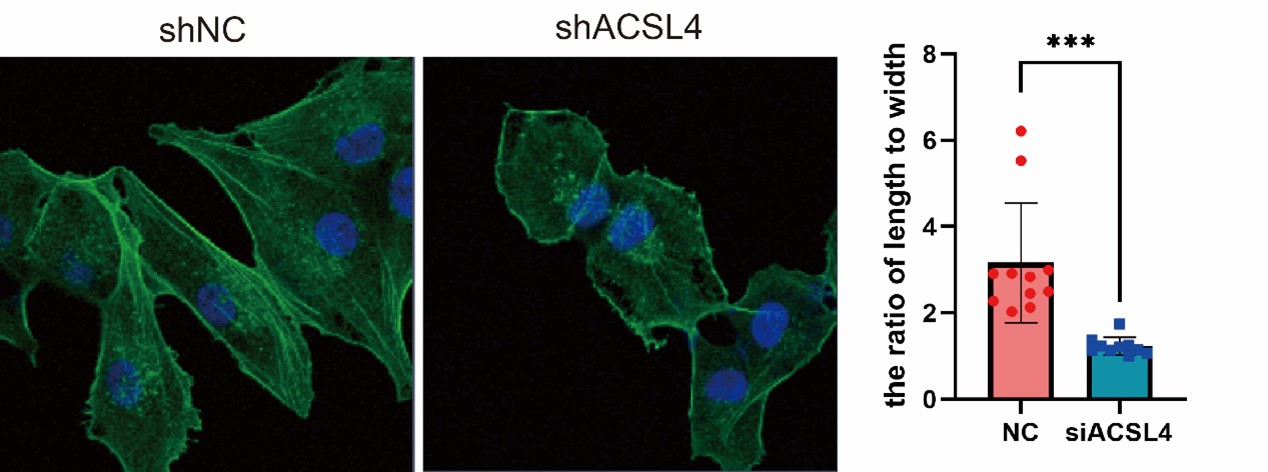
- EMT is characterized by changes in cell morphology, so the staining of cytoskeletons with Phalloidin is needed.
We appreciate the reviewer’s suggestion and performed the staining. The results show that the ACSL4 knockdown cells had a significantly smaller length to width ratio, which indicates the reversion of EMT process, than those of the control group (p<0.05). We have put the results in Supplementary Fig. S4 as follows in Author response image 13:
Author response image 13.
- Additional breast cancer cases or cohorts (such as TMA) should be used to validate the positive correlation between ACSL4 and ZEB2 expression through IHC analysis.
We thank the reviewer for the suggestion. To better understand the positive correlation between ACSL4 and ZEB2 expression, we added more breast cancer cases up to 45 for IHC analysis and validated the positive correlation between ACSL4 and ZEB2. We have put the results into Fig 1 H and I as follows in Author response image 14:
Author response image 14.
Reviewer #3 (Public Review):
The manuscript by Lin et al. reveals a novel positive regulatory loop between ZEB2 and ACSL4, which promotes lipid droplets storage to meet the energy needs of breast cancer metastasis. It is of interest, however, some concerns should be addressed to strengthen the finding.
Major concerns:
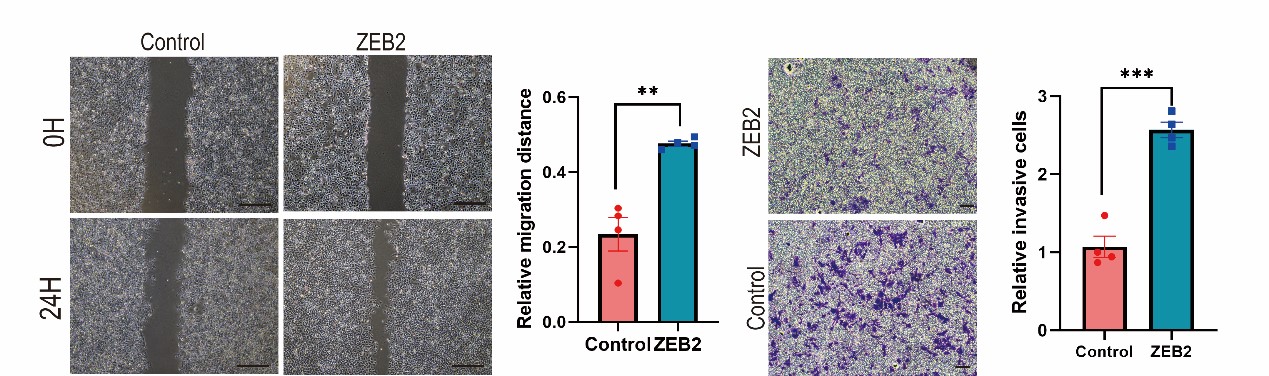
- The effect of ZEB2 overexpression is not fully demonstrated in the whole study. This point should be addressed.
We appreciate the positive comments and constructive suggestion from the reviewer. We have performed ZEB2 over-expressed MCF7 cell line. Over-expression of ZEB2 significantly enhanced the metastatic and invasive capacities of MCF7 cells. (Supplementary Fig. S5A and 5B).
Author response image 15.
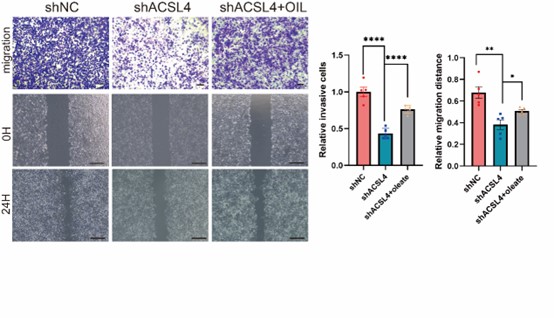
- Does the addition of oleate restore the ability of migration or invasion in ACSL4 knockdown cells?
We thank the reviewer for the question. To address this point, the oleate was added in the culture medium of ACSL4 knockdown cells. As expected, the addition of oleate obviously restores the invasive and metastatic capacities of ACSL4 knockdown cells by 33.12% and 18.61% respectively. We have added the results in the new Fig. 4D as follows in Author response image 16:
Author response image 16.
- Which cellular compartment does ACSL4 localize in and interact with ZEB2 to stabilize ZEB2?
We thank the reviewer for the question. We have repeated the immunofluorescence assay in the MDA-MB-231 cells. We observed that ZEB2 can also be found in the cytoplasm and co-localized with ACSL in some certain regions of the cytoplasm (Supplementary Fig. S11):
- The ubiquitination assay and Co-IP assay are just performed in HEK293T cells. This result should be confirmed in MDA-MB-231 cells or Taxol-resistant MCF-7 cells.
We appreciate the reviewer’s suggestion. We performed the ubiquitination assay and IP assay in MDA-MB-231 cells as follows. The results confirm that knockdown of ACSL4 obviously enhanced the ubiqutination of ZEB2. We have put the results into Fig. 7D and 7F as follows in Author response image 17:
Author response image 17.
- How does ACSL4 regulate ZEB2 at the mRNA level?Please verify.
We thank the reviewer for the thoughtful question. A recent study reported that LD-derived lipolysis provide acetyl-CoA for the epigenetic regulation of gene transcription. We observed that ACSL4 can promote FAO, which generates acetyl-CoA for the epigenetic regulation. It is likely that ACSL4 regulates the ZEB2 mRNA level via lipid-epigenetic reprogramming mechanism, which is worth studying in the future and we had added the following sentences into the second paragraph in the discussion section :
Additionally, we found that ACSL4 promoted ZEB2 transcription as the mRNA level of ZEB2 was significantly reduced after ACSL4 knockdown. A recent study reported that LD-derived lipolysis provide acetyl-CoA for the epigenetic regulation of gene transcription. We observed that ACSL4 can also promote FAO, which can generate acetyl-CoA for the epigenetic regulation. It is likely that ACSL4 regulates the ZEB2 mRNA level via lipid-epigenetic reprogramming mechanism, which is worth studying in the future.
- In Fig. 2F, the silencing efficiency for ACSL4 and ZEB2 should be shown. In addition, the protein level of ZEB2 or ACSL4 in shZEB2 and shZEB2+ACSL4 groups should also be addressed.
We appreciate the reviewer's suggestions. We have added the protein levels in Fig 2F and 2H as follows in Author response image 18:
Author response image 18.
- What is the survival status of patients with both high expression of ACSL4 and ZEB2 in TCGA. In addition, more survival data from databases especially patients with both high expression of ACSL4 and ZEB2 are needed to analyze to support the finding.
We thank the reviewer for the constructive suggestion. We repeated the Kaplan-Meier survival analysis of TCGA RNA-seq data by using R4.3.0 software. The survival data show that the patients with both high expression of ACSL4 and ZEB2 have the worst prognosis in the four groups (P<0.05) ( New Fig. 1D-F).
Reviewer #1 (Recommendations For The Authors):
- Only one siRNA/shRNA was used for knockdown in one cell line. Different siRNAs/shRNAs and multiple cell lines should be used to rule out off-target effects.
We appreciate the reviewer’s suggestion. We have test three siRNA and shRNA for the knockdown efficiency (negative control siRNA or ACSL4 and ZEB2 siRNA were from the company of GenePharma), we chose one sequence with the best knock-down effect.
Author response image 19.
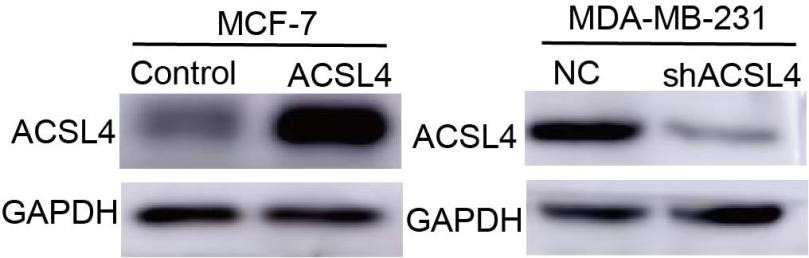
- Western blot data are required to justify the overexpression or knockdown efficiency of ACSL4 in cells in Figure 2 A-C.
We thank the reviewer for the suggestion. we have added the following western blot data in Figure 2:
Author response image 20.
- In Figure 1G, there is a huge variation of the protein input, which makes the results not justified. The authors should repeat the experiments to ensure consistency and reproducibility of the results.
We appreciate the reviewer to point out this problem. Because this is the tissue samples of breast cancer patients. The results are affected by the tumor tissue composition between different patient sample, and it is difficult to obtain fresh tissues. In our paper, paraffin specimens have been used for IHC staining, and the results confirmed that ACSL4 and ZEB2 are positively correlated. We have put the results in the supplementary data.
Reviewer #2 (Recommendations For The Authors):
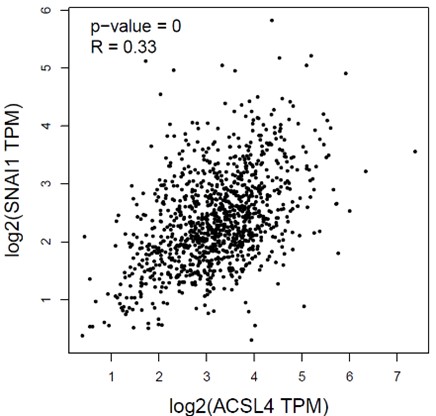
- Data from Figure 1A showed the EMT transcription factor SNAIL was also among the top upregulated genes. Please explain why the association between ACSL4 and ZEB2 was studied instead of ACSL4 and SNAIL.
We appreciate the reviewer’s question. We had calculated the correlation between the ACSL4 and SNAIL by Pearson’s correlation test. The correlation of ACSL4 and SNAIL is 0.33, less than that of ZEB2. Bedsides, the binding motif analysis reveals that the consensus sequence of ZEB transcription family is within the ACSL4 promoter. Thus, we investigated the relationship between ACSL4 and ZEB2 in breast cancer cells.
Author response image 21
- What is the limitation of your study? Please add some relevant description in the part of discussion.
We appreciate the reviewer’s suggestion. We have added the description of limitation of our study in the last paragraph of discussion section as follows:
The limitation of this study is the clinical samples is only 45. The future study should expand the clinical samples and cases to provide more clinical evidence for the crucial role of ACSL4 in breast cancer metastasis.
3). In Figure 3 Figure Legends part, the authors used the word "knockout", which is a description error.
We appreciate the reviewer’s advice. We have corrected "knockout" into "knockdown".
Reviewer #3 (Recommendations For The Authors):
Minor concerns:
- In line 352-353, the statement about whether the high or low expression of ACSL4 and ZEB2 or the advanced breast cancer affects prognosis is inaccurate.
We appreciate the reviewer to point out this problem. We have corrected the statement into “We found that patients with higher ACSL4 or ZEB2 expression, especially those with simultaneous high expression had worse prognosis than those with lower expression ”.
- The title of the seventh part of your results contains a logical error that is opposite to the experimental conclusion.
We truly appreciate the reviewer to point out this problem. We have changed the title of the seventh part of results to “ACSL4 regulates ZEB2 mRNA expression and protein stabilization”.