Peer review process
Revised: This Reviewed Preprint has been revised by the authors in response to the previous round of peer review; the eLife assessment and the public reviews have been updated where necessary by the editors and peer reviewers.
Read more about eLife’s peer review process.Editors
- Reviewing EditorCarlos SimonFoundation Carlos Simon and Valencia University, Valencia, Spain
- Senior EditorWei YanWashington State University, Pullman, United States of America
Reviewer #1 (Public Review):
This study generated 3D cell constructs from endometrial cell mixtures that were seeded in the Matrigel scaffold. The cell assemblies were treated with hormones to induce a "window of implantation" (WOI) state.
The authors did their best to revise their study according to the reviewers' comments. However, the study remains unconvincing and at the same time too dense and not focused enough.
(1) The use of the term organoids is still confusing and should be avoided. Organoids are epithelial tissue-resembling structures. Hence, the multiple-cell aggregates developed here are rather "co-culture models" (or "assembloids"). It is still unexpected (unlikely) that these structures containing epithelial, stromal and immune cells can be robustly passaged in the epithelial growth conditions used. All other research groups developing real organoids from endometrium have shown that only the epithelial compartment remains in culture at passaging (while the stromal compartment is lost). If authors keep to their idea, they should perform scRNA-seq on both early and late (passage 6-10) "organoids". And they should provide details of culturing/passaging/plating etc that are different with other groups and might explain why they keep stromal and immune cells in their culture for such a long time. In other words, they should then in detail compare their method to the standard method of all other researchers in the field, and show the differences in survival and growth of the stromal and immune cells.
(2) The paper is still much too dense, touching upon all kind of conclusions from the manifold bioinformatic analyses. The latter should be much clearer and better described, and then some interesting findings (pathways/genes) should be highlighted without mentioning every single aspect that is observed. The paper needs a lot of editing to better focus and extract take-home messages, not bombing the reader with a mass of pathways, genes etc which makes the manuscript just not readable or 'digest-able'. There is no explanation whatever and no clear rationale why certain genes are included in a list while others are not. There is the impression that mass bioinformatics is applied without enough focus.
(3) The study is much too descriptive and does not show functional validation or exploration (except glycogen production). Some interesting findings extracted from the bioinformatics must be functionally tested.
(4) In contrast to what was found in vivo (Wang et al. 2020), no abrupt change in gene expression pattern is mentioned here from the (early-)secretory to the WoI phase. Should be discussed. Although the bioinformatic analyses point into this direction, there are major concerns which must be solved before the study can provide the needed reliability and credibility for revision.
(5) All data should be benchmarked to the Wang et al 2020 and Garcia-Alonso et al. 2021 papers reporting very detailed scRNA-seq data, and not only the Stephen R. Quake 2020 paper.
(6) Fig. 2B: Vimentin staining is not at all clear. F-actin could be used to show the typical morphology of the stromal cells?
(7) Where does the term "EMT-derived stromal cells" come from? On what basis has this term been coined?
(8) CD44 is shown in Fig. 2D but the text mentions CD45 (line 159)?
(9) All quantification experiments (of stainings etc) should be in detail described how this was done. It looks very difficult (almost not feasible) when looking at the provided pictures to count the stained cells.
(10) Fig. 3C: it is unclear how quantification can be reliably done. Moreover, OLFM4 looks positive in all cells of Ctrl, but authors still see an increase?
(11) Fig. 3F: Met is downregulated which is not in accordance with the mentioned activation of the PI3K-AKT pathway.
(12) Lines 222-226: transcriptome and proteome differences are not significant; so, how meaningful are the results then? Then, it is very hard to conclude an evolution from secretory phase to WoI.
(13) WoI organoids show an increased number of cilia. However, some literature shows the opposite, i.e. less ciliated cells in the endometrial lining at WoI (to keep the embryo in place). How to reconcile?
(14) How are pinopodes distinguished from microvilli? Moreover, Fig. 3 does not show the typical EM structure of cilia.
(15) There is a recently published paper demonstrating another model for implantation. This paper should be referenced as well (Shibata et al. Science Advances, 2024).
(16) Line 78: two groups were the first here (Turco and Borreto) and should both be mentioned.
(17) Line 554: "as an alternative platform" - alternative to what? Authors answer reviewers' comments by just changing one word, but this makes the text odd.
Reviewer #2 (Public Review):
In this research, Zhang et al. have pioneered the creation of an advanced organoid culture designed to emulate the intricate characteristics of endometrial tissue during the crucial Window of Implantation (WOI) phase. Their method involves the incorporation of three distinct hormones into the organoid culture, coupled with additives that replicate the dynamics of the menstrual cycle. Through a series of assays, they underscore the striking parallels between the endometrial tissue present during the WOI and their crafted organoids. Through a comparative analysis involving historical endometrial tissue data and control organoids, they establish a system that exhibits a capacity to simulate the intricate nuances of the WOI.
The authors made a commendable effort to address the majority of the statements. Developing an endometrial organoid culture methodology that mimics the window of implantation is a game-changer for studying the implantation process. However, the authors should strive to enhance the results to demonstrate how different WOI organoids are from SEC organoids, ensuring whether they are worth using in implantation studies, or a proper demonstration using implantation experiments.
Author response:
The following is the authors’ response to the original reviews.
Reviewer #1 (Public Review):
Q1: First of all, the term organoid must be discarded. The authors just seed the endometrial cell mixture which assembles and aggregates into a 3D structure which is then immediately used for analysis. Organoids grow from tissue stem cells and must be passage-able (see their own description in lines 69-71). So, the term organoid must be removed everywhere, to not confuse the organoid field. It is not shown that the whole 3D assembly is passageable, which would be very surprising given the fact that immune and stromal cells do not grow in Matrigel because of the unfavorable growing conditions (which are targeted to epithelial cell growth).
We appreciate for your highlighting concerns regarding our organoid construction.
(1) The organoids in our system were originated from tissue stem cells.
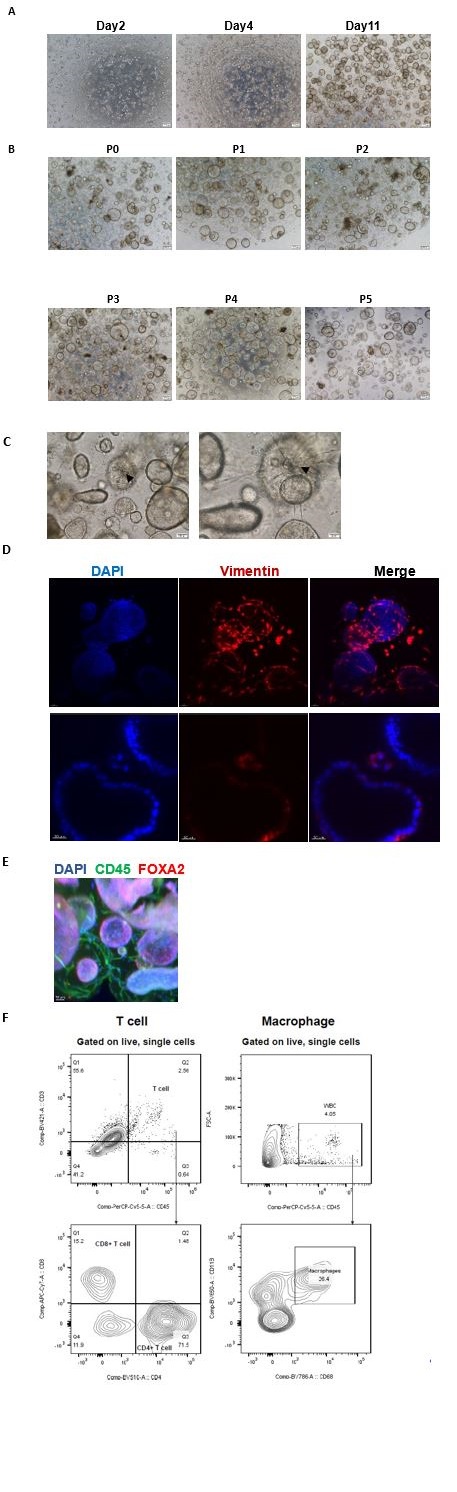
We induced adult stem cells derived from endometrial tissue to construct organoids in vitro by various small molecules (such as Noggin, EGF, FGF2, WNT-3A and R-Spondin1), which involves a complex self-assembly process rather than a mere cellular assembly. Initially, there are single cells and small cell clusters in the system two days after the planting. On the fourth day, the glandular epithelial cells gradually assembled to glands, while the stromal cells spontaneously organized themselves around the glands. On the eleventh day, the endometrial glands enlarged, epithelial cells organized in a paving stone arrangement, and stromal cells established an extensive network. (Author response image1) (Figure 1C)
(2) The organoids we constructed are passage-able.
Most organoids were used for experiments up to the fifth generation, while some are extended to the 10th generation and cryopreserved. (Response Figure 1B, C)
(3) Immune and stromal cells are present in our system from the primary to the fourth generation. In our study, immune and stromal cells were identified not only from scRNA-seq data (third generation of organoids) (Figure 2A), but also from the morphology using 3D transparent staining and light sheet microscopy imaging (third generation of organoids), with Vimentin marking stromal cells, CD45 designating immune cells, and FOXA2 identifying glands. Further, flow cytometric analysis was applied to verify immune cells within the organoids (third generation of organoids). (Response Figure 1D, E, F)
Moreover, Immune cells and stromal cells can grow in Matrigel, which was also found in the study of organoid pioneer Hans Clevers (Hans Clevers et al., Nature Reviews Immunology 2019).
Author response image 1.
(A) The growth condition of endometrial cells was observed from day2 to day11 after plating under an inverted microscope. Scale bar = 200 μm. (B) The endometrial organoids of different passages were observed from P1 to P5. Scale bar = 200 μm. (C) Stromal cells formed an extensive network (down). The arrowhead indicates dendritic stromal cells. Scale bar = 100 μm (left), Scale bar = 50 μm (right). (D) Exhibition of stromal cells marked by vimentin. Nuclei were counterstained with DAPI. The arrow indicates stromal cells. Scale bar = 40 μm (up), Scale bar = 30 μm (down). (E)Exhibition of immune cells marked by CD45 and endometrial gland marked by FOXA2. Nuclei were counterstained with DAPI. The arrow indicates immune cells. Scale bar = 50 μm. (F) Flow cytometric analysis of T cells and macrophages in the endometrial organoid. Gating strategy used for determining white blood cells (CD45+ cells), T cells (CD45+CD3+ cells) and macrophages (CD45+CD68+CD11b+ cells).
Q2: Second, the study remains fully descriptive, bombing the reader with a mass of bioinformatic analyses without clear descriptions and take-home messages. The paper is very dense, meaning readers may give up. Moreover, functional validation, except for morphological and immunostaining analyses (which are posed as "functional" but actually are only again expression) is missing, such as in vivo functionality (after transplantation e.g.) and embryo interaction. Importantly, the 3D structure misses the right architecture with a lining luminal epithelium which is present in the receptive endometrium in vivo and needed as the first contact site with the embryo. So, in contrast to what the authors claim, this is not the best model to study embryo interaction, or the closest model to the in vivo state (line 318, line 326).
Thank you.
(1) We have made the following improvements. Firstly, we have conducted additional experiments to validate the bioinformatics analysis. Secondly, the structure of the manuscript has been refined to ensure logical coherence and clear transitions between paragraphs. Thirdly, important findings have been emphasized to ensure readers’ comprehension and inspiration. Furthermore, the manuscript was revised by both domestic and international experts to enhance the readability and clarity.
(2) For the functional validation, in vivo transfer could not be carried out so far due to ethical limitation. But human embryos are able to develop and grow more efficiently in combining with the receptive endometrial organoids we generated (unpublished data).
(3) As you suggested, we replaced the “closest” with “closer”. It is undeniable that the model cannot completely simulate the in vivo implantation process that the luminal epithelium of the endometrium contacts the embryo first.
Q3: Third, receptive endometrial organoids (assembloids; Rawlings et al., eLife 2021) and receptive organoid-derived "open-faced endometrial layer" (Kagawa et al., Nature 2022) have already been described, which is in contrast to what the authors claim in several places that "they are the first" (e.g. lines 87-88, 316-319, etc). These studies used real organoids to achieve their model (and even showed embryo interaction), while in the present study, different cell types are just seeded and assembled. Hence, logically, immune cells are present which are never found in real organoid models. The only original aspect in the present study is the use of hormones to enhance the WOI phenotype. However, crucial information on this original aspect is missing such as concentration of the hormones, refreshment schedule, all 3 hormones added together or separately, and all 3 required?
Thank you for pointing out these researches referring to endometrial organoids.
(1) While we didn’t explicitly state "the first", we should be careful to use the expressions similar to "the first". It has been changed to a gentle and modest expression, as follows “we are far from understanding how embryo implantation occurs during the WOI due to ethical limitations and fewer in vitro receptive endometrial model” and “which confirms that they are closer to the in vivo state”.
(2) The definition of organoids and the existence of immune cells have been detailed addressed in the first question.
(3) In terms of hormone scheme, hormone concentrations have been detailed in Table S2 of Supplementary. Estrogen was supplemented to the basal medium for the initial two days, after which a combination treatment of MPA, cAMP, PRL, hPL, and HCG was administered for the subsequent six days. The medium was refreshed every two days.
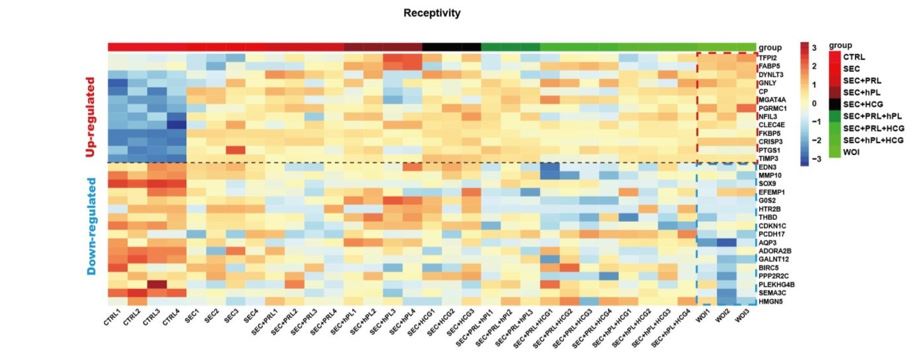
All three hormones were deemed necessary, which was validated by multiple group comparisons. Only the organoids treated with all six hormones together exhibited an endometrial receptivityrelated gene expression profile. (Author response image 2).
Author response image 2.
Heatmap showing receptivity related gene expression profile of organoids in each hormone regimen.
Q4: Moreover, it is not a "robust" model at all as the authors claim, given the variability of the initial cell mixture (varying from patient to patient). Actually, the reproducibility is not shown. The proportions of the different cell types seeded in the Matrigel droplet will be different with every endometrial biopsy. It would be much better to recombine epithelial (passageable) organoids with stromal and immune cells in a quantified, standardized manner to establish a "robust" model.
Thanks for your suggestion.
Firstly, the constructed endometrial organoids generally consist of epithelial, stromal, and immune cells. However, it is undeniable that the cell proportions may vary slightly among different patients. Secondly, the term "robust" is intended to convey strong support for embryo development, which will be supported by our next study (unpublished data). Therefore, robust is replaced here as alternative. Thirdly, as for "reproducibility", the hormone-treated organoids from different women exhibited similarity to the in vivo receptive endometrium through multi-omics analysis, ERT, and various other experiments.
Reviewer #2 (Public Review):
Q1: With endometrial receptivity analysis, they suggest a successful formation of the implantation window in vitro, but this result is difficult to interpret.
Thanks for your question.
We understand that the most effective way to demonstrate endometrial receptivity is embryo implantation, which was conducted simultaneously and will be presented in our next study. In this study, we validated the receptivity based on the current researches.
(1) At the single-cell transcriptome level, the cellular composition and function of the receptive endometrial organoids were similar to those of the in vivo implantation window (Stephen R. Quake et al, 2020).
(2) At the whole organoids level, the receptive endometrial organoids exhibited the similar characteristics in transcriptome and proteome to the in vivo mid-secretory endometrium (Andres Salumets 2017, Qi Yu 2018, Triin Laisk 2018, Edson Guimarães Lo Turco 2018, Xiaoyan Chen 2020, Francisco Domínguez 2020, DavidW. Greening 2021, Norihiro Sugino 2023). The receptive endometrial organoids were also validated by endometrial receptivity test (ERT), which utilized high-throughput sequencing and machine learning to assess endometrial receptivity (Yanping Li et al., 2021).
(3) At the microstructural level under electron microscope, the receptive endometrial organoids exhibited characteristics of the implantation window, such as pinopodes, glycogen particles, microvilli, and cilia.
Overall, the receptive organoids we constructed closely resemble the in vivo implantation window at the single-cell, organoids, and microstructural levels based on existing researches.
Q2: Analyzing transcriptome and proteome information of WOI organoids, authors demonstrate a strong response to estrogen and progesterone, but some comparisons are made with CTRL and SEC, and others only with CTRL, which limits the power of some results. In the same way, some genes related to Cilia and pinopodes appear dominant in WOI organoids, but the comparison by electron microscopy is made only against CTRL organoids.
In subsequent analysis, WOI organoids showed a marked differentiation from proliferative to secretory epithelium, and from proliferative epithelium to EMT-derived stromal cells than SEC organoids. These statements are based on their upregulation of monocarboxylic acid and lipid metabolism, their enhanced peptide metabolism and mitochondrial energy metabolism, or their pseudotime trajectories. However, other analyses (such as the accumulation of secretory epithelium or decreased proliferative epithelium, the increased ciliated epithelium after hormonal treatment, or the presence of EMT-derived stromal cells) show only small differences between SEC and WOI organoids.
Thank you for raising these important questions.
(1) At the organoid level, the differences in transcriptome and proteome between SEC and WOI organoids are not significant. This is understandable because WOI organoids are further induced towards the implantation window based on the secretory phase (i.e. SEC organoids), and both are similar at the overall organoid level.
(2) At the single-cell level, the accumulation of secretory epithelium, decreased proliferative epithelium, increased ciliated epithelium post hormonal treatment, or the presence of EMTderived stromal cells are the fundamental features of the secretory endometrium. Therefore, these features are present in both WOI and SEC organoids. However, the most notable differences lie in the more comprehensive differentiation and varied cellular functions exhibited by WOI organoids compared to SEC organoids.
(3) Regarding electron microscopy, we have now quantitatively compared the presence of various characteristic structures such as microvilli, cilia, pinopodes and glycogen in the CTRL, SEC and WOI groups. It has been observed that WOI organoids possess longer microvilli and increased cilia, glycogen, and pinopodes compared to SEC organoids (Fig2H).
Reviewer #1 (Recommendations For The Authors):
Q1: Several of the key methods are performed by companies, hence not in detail described and therefore not verifiable which is essential for reviewers and readers.
We are grateful for the suggestion. Specific methods have now been incorporated into the "Supporting Information" section. (Line91~102, Line 107~123, Line 132~139)
Q2 - Line 49: It is not shown in the present study whether the WOI organoids are a 'robust' platform.
- Line 76: There is a study (Dolat L., Valdivia RH., Journal of Cell Science, 2021) that developed a co-culture with endometrial organoids and immune cells (neutrophils) which should be mentioned.:
We have reweighed the word and now replace 'robust' with 'alternative' (Line 54). We have considered the reviewer's suggestion and added this citation (Line 82-83) about the cocultivation of immune cells with endothelial organoids, which was not previously cited mainly because the research model was mouse.
Q3: Figure 1: Endometrial organoids possess endometrial morphology and function. - The authors should further explain their decision to add PRL, hCG, and hPL to the organoid culture. Why these particular compounds? What is their specific role during the WOI?
In terms of hormone scheme, estrogen and progesterone promote the transition of endometrial organoids into the secretory phase, and on this basis, pregnancy hormones can further promote their differentiation. PRL promotes immune regulation and angiogenesis during implantation, HCG improves endometrial thickness and receptivity, and HPL promotes the development and function of endometrial glands. Our constructed WOI organoid is in a state conducive to embryo implantation. We aim to develop an in vitro model for embryo implantation study. The detailed explanation of this aspect was initially provided in the Discussion section (Lines 298–313). To enhance the clarity for reviewers and readers regarding the selection of the hormonal regimen, we have now articulated it in the Results section (Lines 124–130).
When selecting hormone formulations, multiple group comparisons were made. It was found that the number, area, and average intensity of organoids in these groups were similar over time. But the WOI organoids showed endometrial receptivity related gene expression profile, which highly expressed genes positively correlated with endometrial receptivity, and lowly expressed genes negatively correlated with receptivity, compared to the other hormone formulations (added to Figure S1E, S1F). Hormone dosage was primarily based on peri-pregnant maternal body or localized endometrium levels (Margherita Y. Turco et al., Nature Cell Biology 2017).
- Line 108: "the endometrial cells" instead of "endometrial organoid"? Because the authors also refer to the stromal cells.
You should be referring to this sentence “The endometrial organoid, consisting of vesicle-like glands, fibrous stromal cells, and other surrounding cells, developed into a 3D structure with the support of Matrigel”. Organoid, a self-assembled 3D structure, consists of multiple cells and closely resembles in vivo tissue or organ. It offers high expansibility, phenotypic, and functional properties. Here, we aim to delineate the endometrial organoid, comprising epithelial cells, stromal cells, and other cellular components that assemble to form intricate 3D structures. Hence, the term "endometrial organoid" is more appropriate.
- Line 110: "the endometrial glands", do the authors mean the endometrial organoids? The authors also mention they enlarge, which must be quantified.
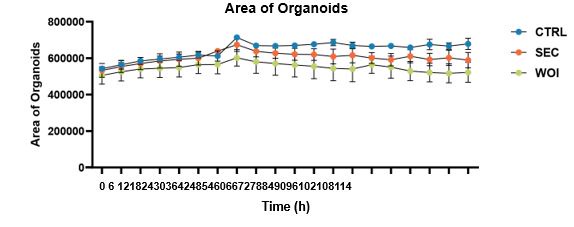
You should be referring to this sentence “As the organoids grew and differentiated, the endometrial glands enlarged, epithelial cells adopted a paving stone arrangement, and stromal cells formed an extensive network”. Here, we mean the “endometrial glands” grow progressively in the organoids. We agree with your suggestion to quantify the change of organoids’ area over time, and found that they increased progressively in all three groups (shown as follows) (Fig.S1E) (Line130-131)
Author response image 3.
The dynamic changes of the area of organoids over time in the CTRL, SEC and WOI organoids.
- Line 112: E-cadherin is a general epithelial marker, not a glandular marker.
We agree with your suggestion and now change to ‘The epithelium marker E-cadherin’ (Line110).
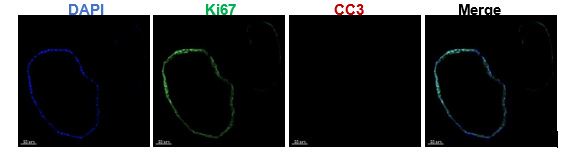
- Line 116: Which group was used for KI67 and CC3 staining?
The CTRL organoids were used for Ki67 and CC3 staining. We have modified this expression in the Figure 1E Legend.
- Line 123: Organoid size (diameter or area) needs to be quantified to claim that WOI organoids grow slower than SEC/CTRL organoids. The same goes for Ki67+ cells for proliferation. In the legend of Fig 1B, the authors in contrast state that the organoids show a similar growth pattern.
We are extremely grateful to you for pointing out this problem. We quantitatively analyzed the size of organoids in the three groups. The area was found to be increasing over time, with the three groups growing the most vigorously in the CTRL group, followed by the SEC group and the WOI group, but the differences were not statistically significant. Relevant results have been added to Figure S1E (Line130-131). There were no significant differences in Ki67 expression of these organoids. Therefore, the three groups of organoids showed a similar growth pattern. We decided to delete the statement “Following hormonal stimulation, WOI organoids exhibited slower growth than SEC and CTRL organoids, while CTRL organoids maintained robust proliferative activity (Fig. 1B)”.
Author response image 4.
The dynamic changes of the area of organoids over time in the CTRL, SEC and WOI organoids.
- Line 126: Fourteen days of organoid treatment is a very long time. Growing organoids may already be dying which should be checked by CC3 staining to prove that organoids are still fully viable.
Endometrial organoids are vigorous in proliferation and have a long survival period due to the presence of adult stem cells. To address your queries effectively, we conducted CC3 staining on the organoids treated for 14 days, revealing negligible expression levels (shown as below).
Author response image 5.
Figure note: The Ki67 and CC3 immunostaining on the organoids after 14-day hormone treatment.
- Line 128: Changes in hormone receptors should be supported by RT-qPCR data to be more convincing
We agree with your suggestion. Here we supplemented the RT-PCR results of hormone receptors as follows (Figure S1D) (Line119-121). PAEP and PGR are associated with progesterone, and OLFM4 and EGR1 are associated with estrogen.
- 1A: Are authors able to see and characterize decidualized stromal cells as indicated in the illustration?
Upon the reviewer's inquiry, we carefully observed the morphology of stromal cells in hormone-treated organoids. Regrettably, the morphology of decidualized stromal cells was not ascertainable through light microscopy in our endometrial organoids.
- 1C: Which treatment condition are the organoids in these images?
This figure showed the bright-field morphology of the CTRL organoids, which is now noted in the Figure 1C legend.
- 1D: PAS staining should be quantified to support the claims.
We agree with your suggestion. The quantitative comparison of PAS staining was conducted in these three groups of organoids (Figure S1G) (Line142-143)
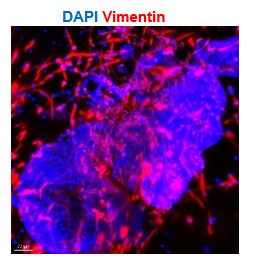
- 1D: Where are the stromal cells in the model? There should be vimentin-positive cells outside of the glands.
The figure 1D illustrates the outcomes of section staining, which owned limitation to displaying stromal cells around the gland. Considering the 3D structure of organoids, we conducted organoid clearing and staining, and observed stromal cells (marked by Vimentin) under light sheet microscope (shown as below). The stromal cells were also presented using this method in the original Figure 2B.
Author response image 6.
Exhibition of stromal cell marked by vimentin of CTRL organoid through whole-mount clearing, immunostaining and light sheet microscopy imaging. Nuclei were counterstained with DAPI. The arrowhead indicates stromal cells. Scale bar = 70 μm.
Figure 2: Developing receptive endometrial organoids in vitro mimicking the implantation window endometrium.
- Line 142: CD44 is not an exclusive marker for immune cells. It has been shown to be expressed in glandular secretory epithelial cells (Fonseca et al., 2023). The authors also mention that CD44 is expressed in stromal cells (line 265). Staining for CD45 (or another immune-specific marker) is needed to demonstrate the presence of immune cells.
We appreciated your suggestions. We demonstrated the distribution of immune cells in organoids using the organoid clearing technique in combination with light-sheet microscopy imaging, using CD45 as a marker (Figure 2C).
- Line 144: What are the proportions of the immune cells? What is the variation between patient samples?
We assessed the proportion of immune cells with the help of flow cytometry and analyzed the proportion of Macrophages and T cells in organoids derived from 8 patients. The proportion of WBC in organoids was about 3%~4% (Figure 2D), among which macrophages were less than 1% and T cells less than 2% (Figure S2E). There existed a very few patients with large heterogeneity, and the proportion of immune cells in most patients was
relatively stable.
- Line 161: What is the endometrial receptivity test (ERT)? Not explained at all.
Endometrial Receptivity Test (ERT) is a kind of gene analysis-based method for detecting endometrial receptivity, which combines high-throughput sequencing and machine learning to analyze the expression of endometrial receptivity-related genes, allowing for a relatively accurate assessment of endometrial receptivity. It is currently used in clinical practice to determine endometrial receptivity and guide personalized embryo transfer (Yanping Li et al., J Transl Med 2021). (line179-183)
- 2A: The authors' dataset is compared to a published dataset. How were they combined? Were they merged, mapped on each other, or integrated? Were all cells employed from the published dataset or specific cell types? Much detail to evaluate the analysis is missing.
We are very grateful for your comments.
(1) The four raw datasets (CTRL, SEC and WOI organoids, and mid-secretory endometrium) underwent batch correction and integration using Harmony. Subsequently, the integrated dataset underwent dimensionality reduction via PCA. The soft k-means clustering algorithm was employed to address batch effects and clustering, utilizing a clustering parameter resolution of 0.5. Finally, the clustering results were visualized using tSNE based on the cell subpopulation classification. (“Methods” Line164-175)
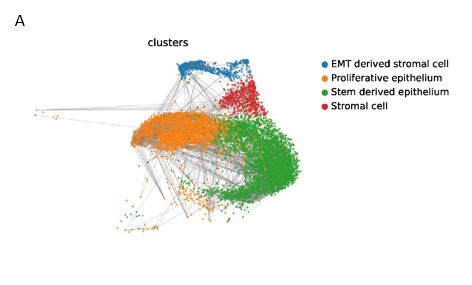
(2) The Figure 2A displayed comparison of glandular and luminal epithelium, secretory epithelium, LGR5 epithelium, EMT-derived stromal cells, ciliated epithelium, and glandular secretory epithelium (shown as Figure S2C~S2D) (Line150-154)
- 2E: Please add the cell type names above the heatmaps to improve readability.
Thanks to your suggestion, we have added the cell type names above the heatmaps.
- 2G: The difference between the left and right graphs is not clear from the figure itself. Improve by adding a title and more explanation.
Thanks for your careful review. We have added the title to the left and right graphs.
Supplementary Figure 3 is referenced with Figure 2. Supplementary Figure 2 is referenced with Figure 3. The order needs to be changed.
Thanks for your careful review. We have changed the order.
- S3B: Typical markers for annotation of the different cell clusters are not included and therefore it is not convincing enough that annotations are correct. E.g. Epithelial markers (EPCAM, CDH1), Stromal cells (VIM, PDGFRA), SOX9+LGR5+ cells (SOX9, LGR5). How were the EMT-derived stromal cells designated? It is not clear from the data whether they are in fact EMT-derived or whether they show epithelial markers as well (stated in line 246).
We deeply appreciate your suggestion. We provided more details to describe the cell clustering as the following. Single-cell transcriptomics analysis referred to CellMarker, PanglaoDB, Human Cell Atlas, Human Cell Landscape, and scRNASeqDB, and previous endometrium related studies. (W. Wang et al., Nat Med 2020, P. D. Harriet C. Fitzgerald et al., PNAS 2019, K. M. Thomas, M Rawlings et al., eLife 2021, L. Garcia-Alonso et al., Nat Genet 2021)
(1) SOX9+LGR5+ cells: SOX9 and LGR5 are both proliferative markers. SOX9 is expressed in all clusters dispersedly. LGR5 is mainly expressed in two clusters, one of which is stem derived epithelium, and the other cluster expresses LGR5 in a scattered manner. Refer to the markers of SOX9+LGR5+ cells, SOX9+LGR5- cells, and SOX9+ proliferative cells in 2021 Nature Genetics (L. Garcia-Alonso et al., Nat Genet 2021), the cells in this cluster expressed high levels of NUAK2, CNKSR3, FOS and LIF, which was consistent with the expression profiles of SOX9+LGR5+ cells and SOX9+ proliferative cells. However, considering that the number of cells expressing LGR5 was relatively small, this cluster of cells was renamed SOX9+ proliferative epithelium.
Figure 3: Receptive endometrial organoids recapitulate WOI-associated biological characteristics. - Line 173-174: The WOI organoids should be compared in detail to the SEC organoids in addition to the CTRL organoids, to show that this WOI model and new hormonal treatment is providing better results compared to the SEC organoids and the results obtained in previous studies.
Thanks for your suggestion. At the organoid level, the differences in transcriptome and proteome between SEC and WOI organoids are not significant. This is understandable because WOI organoids are further induced towards the implantation window based on the secretory phase (i.e. SEC organoids), which prompted us to continue exploring at the single-cell level.
- Line 190: Quantification of pinopodes is required to claim that they are more densely arranged in WOI organoids.
- Line 190-191: Again, is there a difference in pinopode presence between the WOI and SEC organoids to show that the WOI organoids are really distinct and a better model?
We agree with the reviewer’s suggestion and quantified the pinopodes. The CTRL, SEC and WOI organoids were found to have increasing numbers of pinopodes, with WOI organoid owning the most abundant pinopodes under electron microscope. (Figure 2H) (Line184-186)
- Line 194: Also here, quantification of the glycogen particles is missing.
We agree with your suggestion. We have quantified the area of glycogen particles under electron microscope in the CTRL, SEC and WOI organoids. It was found that WOI organoid had the most glycogen particles. (Figure 2H) (Line184-186)
- 3C: There is no difference between SEC and WOI organoids condition for OLFM4 and PRA/B. What is the purpose then of adding extra hormones if no difference is present?
The figure 3C indicated that there was no significant difference in OLFM4 and PRA/B level (reflecting estrogen and progesterone responsiveness) in SEC and WOI organoids at the organoids level. It is understandable because WOI organoids are induced further into the implantation window on the basis of the secretory phase (i.e., SEC organoids), and both are similar at the overall level of organoids. Based on this, we further explored the differences between WOI organoids and SEC organoids at the single-cell level.
- 3G: A higher magnification is necessary to evaluate cilia staining. From these images, it seems like CTRL organoids also express acetyl-a-tubulin.
Thanks for your suggestion. The figure has been enlarged and shown as below. The acetyl-a-tubulin of WOI organoids is different from that of CTRL organoids in morphology and expression level. The glands of WOI organoids have small green tips (expressing acetyl-α-tubulin) convex toward the lumen. WOI organoids expressed higher level of acetyl-α-tubulin than CTRL organoids. (Now replaced with Figure 3G in the revised draft).
Figure 4: Structural cells construct WOI with functionally dynamic changes
- Line 211: To which figure are these claims referring to?
You should be referring to this sentence “In terms of energy metabolism, the WOI organoids exhibited upregulation of monocarboxylic acid and lipid metabolism, and hypoxia response”. Up-regulation of monocarboxylic acid and lipid metabolism in WOI organoids is reflected in Figure 3B, and up-regulation of hypoxia responses is reflected in Figure S3F.
- In general, it should be stated in the text that CellPhoneDB is a useful tool to investigate ligandreceptor interactions, however, it only proposes potential interactions. To validate such interactions, stainings and functional assays are required.
Thanks for your suggestion. The CellphoneDB was briefly introduced in the "Methods" section of "Supporting information" originally. Now it has been explained in the line 256-257 of main text.
We agree that staining and functional assays are required to validate the ligand-receptor interactions. Therefore, we used the proximity ligation assay (PLA) to verify the trend of interaction. (Figure S2J, Line259-261, Line 277-279, Line 285-288)
- Line 243: Please describe the process of EMT in the endometrium more specifically.
EMT is a common and crucial biological event in the endometrium during the implantation window. During the EMT process, epithelial cells lose their epithelial characteristics while gaining migratory and invasive properties of fibroblasts.
During the attachment and adhesion phases of embryo implantation, interaction mediated by trophoblastic factors (e.g. integrins) and maternal ECM factors (e.g. fibronectin) induce the eventual EMT in the trophectoderm. During the peri-implantation period, microRNAs, (e.g. miR429 and miR-126a-3p) which regulate EMT, are expressed in the maternal luminal epithelium to different degrees, mediating its transformation process as the blastocyst invades the maternal decidua. The epithelium of endometrium transforms to epithelioid stromal cells with increased migratory and invasive capacities through the EMT process. The decidual stromal cells migrate away from the implantation site, having acquired increased motility. (Line 265-267)
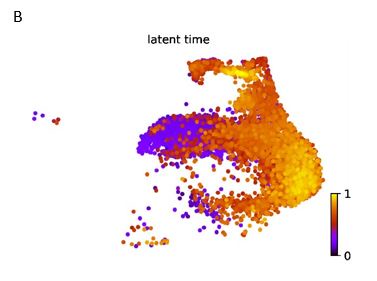
- Lines 247-251 and 313-316: the claim that proliferative epithelium transforms into EMT derived stromal cells by pseudotime trajectory is too bold and must be underpinned by other means. Pseudotime analysis only suggests and is by definition biased since the first/originating population must be defined by the operator.
In addition to pseudotime analysis based on monocle, RNA rate analysis based on scVelo is also used for cell evolution analysis. They can prove each other if both analyses indicate the transformation from proliferative epithelium to EMT-derived stromal cell. RNA rate analysis automatically determines the direction of differentiation, which can be used as evidence to determine the starting point of pseudotime analysis.
RNA rate analysis showed that the EMT derived stromal cell was most closely connected to the proliferative epithelium. Besides, the pseudotime point plot inferred that the proliferative epithelium was the root cell. It can be mutually proved with pseudotime analysis that the transformation from proliferative epithelium to EMT-derived stromal cell.
Author response image 7.
RNA rate junction diagram (To infer intercellular connectivity)
Author response image 8.
Time differentiation of cells
Discussion
- Line 300-302: It would be interesting to investigate ATP production and IL8 release in the WOI organoids to validate with findings from in vivo.
To answer this point of your interest, we purposely examined ATP production and IL8 release. It was found that WOI organoids indeed produced much more ATP and IL8 than CTRL and
SEC organoids (Figure S3L) (Line323-324)
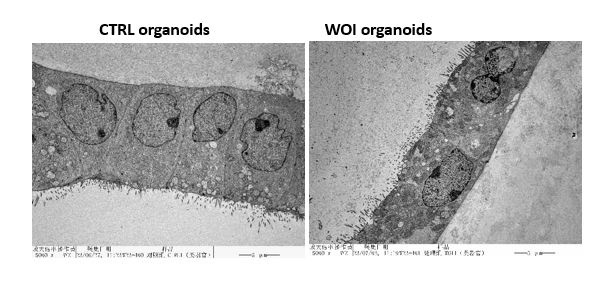
- Line 313-316: Do the WOI organoids lose polarity and cell-to-cell junctions?
Transcriptome sequencing revealed downregulation of cell adhesion and RHO GTPase signaling in WOI organoids (Figure 3B). Electron microscopy revealed that the cellular arrangement of WOI organoids was slightly looser than that of CTRL organoids, but the microvilli were still oriented toward the medial side of the glands and did not undergo polarity reversal (shown as below).
Author response image 9.
Electron micrograph of the CTRL (left), and WOI (right) endometrial organoid. Scale bar = 5 μm.
- Line 322: Where is the data that shows that 'a decreased abundance of immune cells', is observed?
A decreased abundance of immune cells was observed through single-cell transcriptome sequencing and flow cytometry. The number of immune cells was reduced in WOI organoids compared to CTRL organoids in single-cell sequencing results (Figure 4A). Besides, flow cytometry also showed that the percentage of WBCs in WOI organoids was lower than that in CTRL organoids (Figure S2F).
- Line 324: Elaborate more on how the immune cell composition differs from the endometrium.
The differences of immune cell composition between organoids and endometrium were mainly reflected in the proportion of WBC, the proportion of immune cell subtypes and the changes of T cells after entering the implantation window.
Firstly, the proportion of WBCs in organoids was lower than that in endometrium. Flow cytometry showed that the proportion of WBC in organoids was about 3%~4% (Figure 2D), but the proportion of WBCs in endometrium was about 8% (W. Wang et al., Nat Med 2020). Secondly, the proportions of T cells and macrophages in organoids were about 2%~3% and 1% (Figure 2D), respectively, but the proportions of lymphocytes and macrophages in endometrium were 7%~8% and 0.6%~0.7% (W. Wang et al., Nat Med 2020). Besides, after entering the implantation window, T cells in WOI organoids decreased (Figure S2F), while T cells in endometrium increased (W. Wang et al., Nat Med 2020). These three aspects have differences in vivo and in vitro. (Line347353)
Material and Methods
- What are the concentrations of all medium components?
Thanks to your suggestions. The concentrations of all medium components have now been refined in Table S1.
- Authors mention 10x while Smartseq2 is mentioned in Dataset S7?
Thanks for your careful review. Single cell transcriptome sequencing in this study was done using 10X Genomics. Smartseq2 was used to sequence the transcriptome of a gland and its surrounding cells, which can be regarded as small bulk RNA sequencing. A small number of cells are utilized in Smartseq2 to construct a full-length mRNA library with enhanced transcript sequencing coverage, making it particularly well-suited for small-scale samples such as organoids.
The data in Dataset S7 are acquired from small bulk RNA-seq with Smartseq2.
Reviewer #2 (Recommendations For The Authors):
Q1: The theoretical choice of extra reagents added to the WOI organoids culture (PRL, hCG, and hPL) is theoretically justified, but not experimentally. On what previous studies, or performed experiments, are the choice of conditions used based?
When selecting hormone formulations, multiple group comparisons were made. It was found that the number, area, and average intensity of organoids in these groups were similar over time. But the WOI organoids showed endometrial receptivity related gene expression profile, which highly expressed genes positively correlated with endometrial receptivity, and lowly expressed genes negatively correlated with receptivity, compared to the other hormone formulations (added to Figure S1E, S1F). Hormone dosage was primarily based on peri-pregnant maternal body or localized endometrium levels (Margherita Y. Turco et al., Nature Cell Biology 2017).
Q2: Text in line 111 indicates that "stromal cells formed an extensive network", but vimentin fluorescence is not present on any image surrounding organoids in that figure. This assertion could only be supported by the subsequent results in Figure 2B. In addition, it is not indicated what kind of organoids have been used for these experiments
The stromal cells arranged around the glands in the 3D structure (as shown in Figure 1C and Figure 2B), where bright-field high magnification photography, clearing staining of the organoids, and light microscopy imaging were used, respectively. However, there are many steps of fixation, embedding, staining and elution during the immunostaining of sections. It is difficult to preserve the arrangement and morphology of the stromal cells in the slice, so the stromal cells were not intentionally captured in the other images.
Figure 1C and Figure 2B are both CTRL organoids, which are now noted in the corresponding figure legend section.
Q3: It is not clear how glycogen secretion into the lumen is assessed in Figure 1D.
Glycogen from the subnuclear region of the glandular cells gradually reaches the top of the cells, i.e., the supranuclear region, and is discharged into the glandular lumen as parietal plasma secretion. Glycogen-containing eosinophilic secretion can be seen in the glandular lumen in Figure1D.
Q4: Assertions about differences in proliferation between groups are purely subjective; some kind of measurement and analysis would be necessary to be sure that there is differential proliferation based on Figure 1B.
We are extremely grateful to you for pointing out this problem. We quantitatively analyzed the size of organoids in the three groups. The area was found to be increasing over time, with the three groups growing the most vigorously in the CTRL group, followed by the SEC group and the WOI group, but the differences were not statistically significant. Relevant results have been added to Figure S1E (Line130-131).
Q5: For progesterone receptor expression analysis organoids are cultured for fourteen days. What is the basis for this change in culture time?
The choice of time point here is based on the secretary period of 14 days in the female menstrual cycle, when the endometrium is stimulated by estrogen and progesterone to maximized
level.
Q6: "n" number of individuals analysed through single-cell transcriptomics is not indicated.
One patient's endometrium was simultaneously constructed into CTRL, SEC and WOI organoids, which were then subjected to single-cell transcriptome sequencing. This is described in the Supporting Information (Line 141-142).
Q7: Where does the classification of EMT-derived stromal cells come from?
EMT is a common and crucial biological event in the endometrium during the implantation window. During the EMT process, epithelial cells lose their epithelial characteristics while gaining migratory and invasive properties of fibroblasts.
This cluster of cells expresses both epithelium markers CDH1 and EPCAM, and specifically expresses high levels of the EMT-related stromal cell markers AURKB, HJURP and UBE2C. During endometrial EMT, AURKB upregulates MMP2, VEGFA/Akt/mTOR and Wnt/β-catenin/Myc pathways to induce EMT (Zhen Wang et al., Cancer Manag Res 2020). HJURP also activates Wnt/β-catenin signaling to promote EMT (Y Wei et al., Eur Rev Med Pharmacol Sci 2019, Tianchi Chen et al., Int J Biol Sci 2019). UBE2C is upregulated by estrogen to promote EMT (Yan Liu et al., Mol Cancer Res 2020). Therefore, this cluster was defined as "EMT-derived stromal cells”.
Q8: In the endometrial receptivity test (ERT), endometrium sample data matches with prereceptive endometrium and WOI organoids data matches with a receptive endometrium, but why there is no information about CTRL and SEC organoids?
We performed ERT on these samples at a time when our hospital has a cooperative project with Yikon Genomics (Jiangsu, China). However, only endometrium and WOI organoids were sent for testing due to the limited quotas. Considering the end of cooperation and batch effect, no more CTRL and SEC organoids were tested. Moreover, the current ERT is a machine learning model based on the sequencing data of endometrium samples. But there are still differences in cellular composition between endometrial organoids and endometrium. Thus, the results need to be interpreted in conjunction with other results.
Q9: When analysing the transcriptome and proteome, some comparisons are made between WOI vs CTRL and SEC, or just WOI vs CTRL. It would be interesting to have all the comparisons since the power of WOI organoids lies in their differences with SEC organoids.
Thanks for your suggestion. At the organoid level, the differences in transcriptome and proteome between SEC and WOI organoids are not significant. This is understandable because WOI organoids are further induced towards the implantation window based on the secretory phase (i.e. SEC organoids), which prompted us to continue exploring at the single-cell level.
Q10: Electron microscopy comparisons with respect to pinopods, cilia, and microvilli are only performed between WOI and CTRL. It would be interesting to check it with SEC.
We now quantitatively compared the presence of various characteristic structure like microvilli, cilia, pinopodes and glycogen in the CTRL, SEC and WOI organoids. It was found that WOI organoid had longer microvilli and increased cilia, glycogen, and pinopodes (Figure 2H).
Q11: Line 190 states that pinopods are arranged more densely in WOI organoids than in CTRL organoids. Seems to be a subjective observation. Is there an objective method to quantify this?
We agree with the reviewer’s suggestion and quantified the pinopodes. The CTRL, SEC and WOI organoids were found to have increasing numbers of pinopodes, with WOI organoid owning the most abundant pinopodes. (Figure 2H) (Line184-186)
Q12: Some characteristics are very similar between WOI and SEC organoids (such as the accumulation of secretory epithelium or decreased proliferative epithelium, the increased ciliated epithelium after hormonal treatment, or the presence of EMT-derived stromal cells). The authors should complement the discussion by objectively justifying the use of WOI versus SEC organoids. Would they be useful in more specific cases or at a general level when studying implementation?
Thanks for your comments. WOI organoids are differentiated from SEC organoids towards the implantation window. Therefore, WOI organoids are suitable for studying periimplantation physiological changes or exploring pathological mechanisms. SEC organoids can be used when studying only a range of pathological problems such as endometrial secretory phase changes or hormone reactivity. (Line 365-368)
Q13:ExM media is described in Table S1, but it does not include the concentration of the different reagents in the culture medium, which is the most interesting data about the ExM medium.
Thanks to your suggestions. The concentrations of all medium components have now been refined in Table S1.
Q14: It is not specified which organoid pass is used in each experiment. Is it always the same pass?
Our experiments were conducted using P1~P3 generation endometrial organoids, as specified in the “Supporting Information” Line 54~55.
Q15: As a protocol for freezing organoids is included in materials and methods, do the authors use freshly cultured organoids or do they cryopreserve them and thaw them for culturing?
Thanks for your question. We used freshly cultured organoids in the manuscript. We listed the freezing protocol to illustrate that the constructed organoids can be frozen and recovered for special experimental needs and the establishment of sample banks.
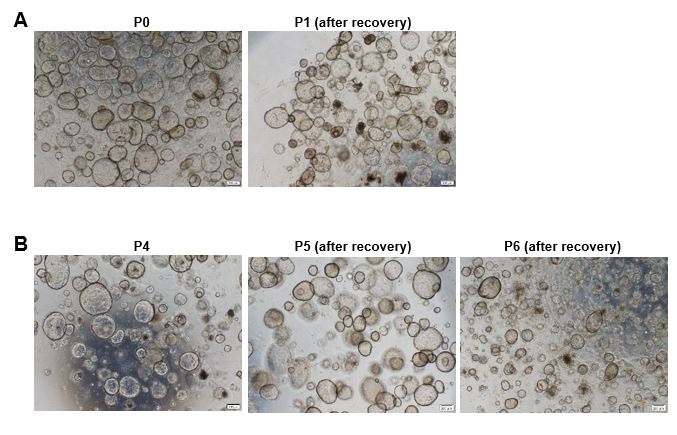
Q16: The most important point: Neither of the two studies that developed human endometrial organoids from tissue biopsies (Boretto et al. 2017 and Turco et al. 2017), observed stromal cell growth in culture. They disappeared between the first and second pass (as indicated by Turco et al. 2017). How do the authors justify the presence of stromal cells in their organoid culture if they rely on the protocols previously described by these research groups? If it is the case that they can only use the initial pass (freshly planted cells from endometrium), it does not make sense to include the freezing of the different passes in materials and methods, since the expansion capacity of the culture would be lost, which implies a major limitation of the model.
Thanks for your question.
(1) We did not completely follow the protocols of these research groups. To maximize the recovery of both epithelial and stromal cells, we optimized key steps such as tissue digestion and cell strainer filtration. We shortened the digestion time to 20 minutes to protect cells from the digestion solution and retain some cell aggregates, which are beneficial for maintaining cell stemness and preserving stromal and immune cells cluster. The 40 μm filter membrane was used to isolate the endometrial cells, which may acquire both epithelial, and stromal cells.
(2) Our experiments were conducted using P1~P3 generation of freshly constructed organoids. However, we also used recovered organoids when fresh endometrial samples were not available due to the COVID-19 epidemic. It was found that the organoids (e.g., P0~P5) still exhibited vigorous growth condition after recovery and could continue to be cultured by passaging (shown as below).
The recovered organoids can be used for special experiments and biobank establishment.
Author response image 10.
The endometrial organoids of different passages were observed before cryopreservation and after recovery. Scale bar = 200 μm.
Q17: It is not clear which organoids include Figure S2F. Does it include the three types of organoids or just WOI organoids?
This circle diagram showed the functions of upregulated genes in the WOI group compared to CTRL group from combined transcriptome and proteome analysis, which has been labeled in the figure legend section.