Peer review process
Revised: This Reviewed Preprint has been revised by the authors in response to the previous round of peer review; the eLife assessment and the public reviews have been updated where necessary by the editors and peer reviewers.
Read more about eLife’s peer review process.Editors
- Reviewing EditorVolker DötschGoethe University Frankfurt, Frankfurt am Main, Germany
- Senior EditorVolker DötschGoethe University Frankfurt, Frankfurt am Main, Germany
Reviewer #1 (Public Review):
Summary:
This technical report by Kugler at al., expands the application of a fluorescence-based reporter to study the conformational state of various kinases. This reporter, named KinCon (Kinase Conformation), interrogates the conformational state of a kinase (i.e., active vs. inactive) based on engineering complementary fusion proteins that fluoresce upon interaction. This assay has several advantages as it allows studying full-length kinases, that is, the kinase domain and regulatory domains, inside the cell and under various experimental conditions such as the presence of inhibitors or activator proteins, and in wildtype and mutants involved in disease states.
Strengths:
One major strength of this study is that it is quite comprehensive. The authors use KinCon for four different kinases, BRAF, LKB1, RIP and CDK4/6. These kinases have very different regulatory elements and associated proteins, which the authors explore to study their conformational state. Moreover, they use small molecule inhibitors or mutations to further dissect how the conformational state of the kinase in disease states. The collective set of results strongly suggests that KinCon is a versatile tool that can be used to study many kinases of biomedical and fundamental importance. Given that kinases are extensively studied by researchers in academia or industry, KinCon could have a broad impact as well.
Weaknesses:
This manuscript, however, also has several weaknesses that I outline below. These weaknesses decrease the overall level of impact on the manuscript, as is.
• The manuscript is exceedingly long. For instance, the introduction provides background information for each kinase that is further expanded in the results section. I think the background information for each kinase in the Introduction and Results sections can be significantly reduced to highlight the major points. Otherwise, not only does the manuscript become too long, but also the main points get diluted.
• Similarly, the figure legends are very long, providing information that is already in the main text or in Methods. The authors should provide the essential information to understand the figure.
• A major concern throughout the manuscript is the use of the word "dynamics," which is used in the text in various contexts. The authors should clarify what they understand for dynamics of conformation. Are they measuring how the time-dependent process by which the kinase is interconverting between active and inactive states? It seems to me that the assays in this report evaluate a population of kinases that are in an open or close conformation (i.e., a particular state in each experimental condition) but there is not direct information how the kinase goes from one state to the other. In that sense, the use of dynamics is unclear. Also, the use of dynamics in different sentences in ambiguous. Here are a few examples but this should be revised throughout the manuscript:
- Line 27: dynamics of full-length protein kinases. Is this referred to dynamics of conformational interconversion between inactive and active states?
- Line 138: dynamic functioning of kinases. No clear what that means.
- Line 276: ... alters KinCon dynamics. Not clear if they are measuring time-dependent process or a single point.
- Figure legend 4F: dynamics of CDK4/6 reporters. Again, not clear how the assay is measuring dynamics.
Nonetheless, in my opinion the authors use proper terminology that describes their assay in which the term dynamics is not used: Title (... impact of protein and small molecule interactions on kinase conformations) and Line 89 (... reporter can be used to track conformational changes of kinases...)
• The authors use the phrase that KinCon has predictive capabilities (abstract and line 142). What do the authors refer to this?
• The authors indicate that KinCon is a highly sensitive assay. Can the authors elaborate on what high sensitivity means? For example, can they discuss how other fluorescence-based approaches that are less sensitive would not be able to accomplish the same type of results or derive similar conclusions? Can they provide a resolution metric both in space and time? Given that the authors state that this is a technical report, this information is of relevance.
• The authors nicely describe how KinCon works in Figure 1B and part of 1C. I do think that the bottom of panel 1C needs to be revised, as well as the text describing the potential scenarios of potency, efficacy and synergism.
- One issue with this part of Figure 1C is that it is not clear what the x-axis in the 3 plots refer to. Is this time? Is this concentration of a small molecule, inhibitor or binding partner? This was confusing also in the context of the term dynamics used throughout the text. The terms potency, efficacy and synergism should be subtitles or the panels and the x-axis should be better defined, especially for a non-specialized reader.
- Related to this part of Figure 1C is the text. The authors mention potency, effectiveness and synergy (Line 195). Can the authors use more fundamental terminology related to these three scenarios, for example, changes in activation constant, percent of protein activates? Also, why synergy is only related to effectiveness? Can synergy also be associated to potency?
- Lastly, the use of these three cartoons gives the impression that the experimental results to come will follow a similar representation. Instead, the results are presented in bar plots for many different conditions. I think this will lead to confusion for a broad audience.
• For a non-expert reader, can the authors clarify the use of tracking basal conformations vs. transient over-expression of the various KinCon constructs? Moreover, the authors use the term transient over-expression for 10, 16, 24 and 48 h (Line 203). This, to a non-expert reader, seems not transient.
• Regarding Figure 1E and similar graphical representations: Why is the signal (RLU) non-linear with time? If the fluorescence of the KinCon construct is linearly related with its expression or concentration inside the cell, one would expect a linear increase. Have the authors plotted RLU/Expression band intensity to account for changes in protein concentration? For instance, some of the results within Figure 3 are normalized to concentration on the reporter expression level.
• For the results with LKB1, the authors claim that intermediate fold change in fluorescence (Figure 2E) is due to a partially closed intermediate state (Line 262). Can the authors discard the possibility by which there is a change in populations of active and inactive that on average give intermediate values?
• The authors claim in Line 274 that mutations located at the interface of the LKB1/STRADalkpha complex affect interactions and hypothesize that allosteric communication between LKB1 and STRADalpha is essential for function. Given that this mutations are at the interaction interface, why would the authors postulate an allosteric mechanism that evokes an effect distant to the interaction/active site? Could it be that function requires surface contacts alone that are disrupted by the mutations?
• I was unable to find text to explain the following: Figure 2I shows the mutation R74A as n.s., but in the text only W308C is mentioned to not change fluorescence. Could the authors clarify why R74A is not discussed in the text? Maybe this reviewer missed the text in which it was discussed. Similarly, the author states in line 326 that the study included an analysis of RIPK2. However, I was unable to find results, graphs or additional text discussing RIPK2.
• Some figures of RLU use absolute values, percentages and fold change. Is there a reason why the authors use different Y-axis values? These should be explained and justified in Methods. Similarly, bars for wt in Figures 3D, G, or 4D, E,F show no errors. How are the authors normalizing the data and repeats so that there is no error, and are they treating the rest of the data (i.e., mutants and/or treated with small molecules) in the same way?
• Lastly, the section starting in Line 472 reads more like a discussion of results from different type of inhibitors used in this study that results on its own. The authors should consider a new subtitle as results or make this section a discussion.
Reviewer #2 (Public Review):
Summary:
Protein kinases have been very successfully targeted with small molecules for several decades, with many compounds (including clinical drugs) bringing about conformational changes that are also relevant to broader interactions with the cellular signaling networks that they control. The authors set out to develop a targeted biosensor approach to evaluate distinct kinase conformations in cells for multiple kinases in the context of incoming signals, other proteins and small molecule binding, with a broad goal of using the KinCon assay to confirm (and perhaps predict) how drug binding or signal perception changes conformations and outputs in the presence of cellular complexes; this work will likely impact on the field with cellular reporters of kinase conformations a useful addition to the toolbox.
Strengths:
The KinCon reporter platform has previously been validated for well-known kinases; in this study, the team evaluate how to employ a full-length kinase (often containing a known pathological mutation). The sensitive detection method is based on a Renilla luciferase (RLuc)protein fragment complementation assay, where individual RLuc fragments are present at the N and the C terminus of the kinase. This report, which is both technical and practical in nature, co-expresses the kinase with known interactors (at low levels) in a high throughput format and then performs pharmacological evaluation with known small molecule kinase modulators. This is explained nicely in Figure 1, as are the signaling pathways that are being evaluated. Data demonstrate that V600E BRAF iexposed to vemurafenib is converted to the inactive conformation, as expected. In contrast, the more closed STRAD𝛼 and LKB1 KinCon conformations appear to represent the more active state of the complexed kinase, and a W308C mutation (evaluated alongside others) reverses this effect. The authors then evaluated necroptotic signaling in the context of RIPK1/3 under conditions where RIPK1 and RIPK3 are active, confirming that the reporters highlight the active states of both kinases. Exposure to compounds that are known to engage with the RIPK1 arm of the pathway induce bioluminescence changes consistent with the opening (inactivation) of the kinase. Finally, the authors move to an important drug target for which clinical drugs have arrived relatively recently; the CDK4/6 complexes. These are of additional importance because kinase-independent functions also exist for CDK6, and the effects of drugs in cells usually relies on a downstream marker, rather than demonstration of direct protein complex engagement. The data presented are interpreted as the formation of complexes with the CDK inhibitor p16INK4a; reducing the affinity of the interaction through mutations drives an inactive conformation, whilst the application of CDK4/6 inhibitors does not, implying binding to the active conformation.
Weaknesses:
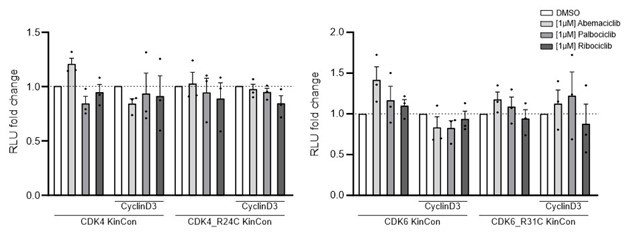
(1) The work is very solid, and uses examples from the literature and also extends into new experimental space. An obvious weakness is mentioned by the authors for the CKDK data, in that measurements with Cyclin D (the activating subunit) are not characterised, although Cyclin D might be assumed to be present?
(2) The work with the trimeric LKB1 complex involves pseudokinase, STRADalpha, whose conformation is also examined as a function of LKB1 status; since STRAD is an activator of LKB1, a future goal should be the evaluation of the complex in the presence of STRAD inhibitory/activating small molecules.