APP interacts with LRP4 and agrin to coordinate the development of the neuromuscular junction in mice
Figures
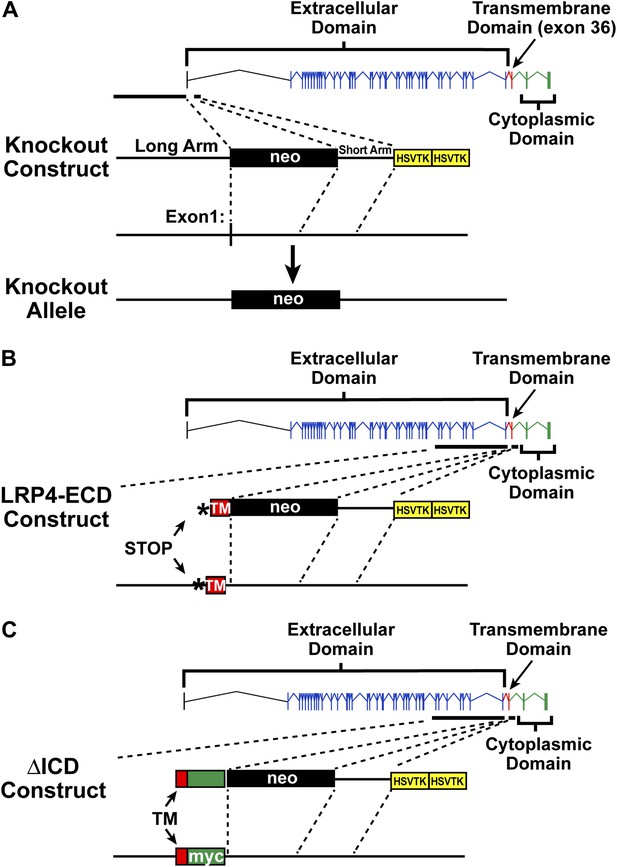
Targeting vectors.
Diagrammatic representation of gene replacement strategies used to generate Lrp4−/− (null allele, A), Lrp4ECD/ECD (hypomorphic allele without membrane anchor, B), and Lrp4ΔICD/ΔICD (C) mice. Exons encoding the LRP4 ECD are depicted in blue and indicated by brackets. The exon encoding the transmembrane segment is shown in red, cytoplasmic domain encoding exons in green. To generate the Lrp4 null allele, the transcription start site and exon 1 were replaced with a neo cassette (A). To generate the Lrp4 ΔICD allele, a cDNA cassette encoding the transmembrane segment of LRP4 (TM, red) followed by a Myc epitope and bovine growth hormone 3′UTR was introduced into the targeting vector described by Johnson et al. (2005). This results in the normal expression of the LRP4 ECD and transmembrane segment, but complete replacement of the ICD with a Myc tag (C). The generation of the LRP4 ECD allele has been described in Johnson et al. (2005) (B).
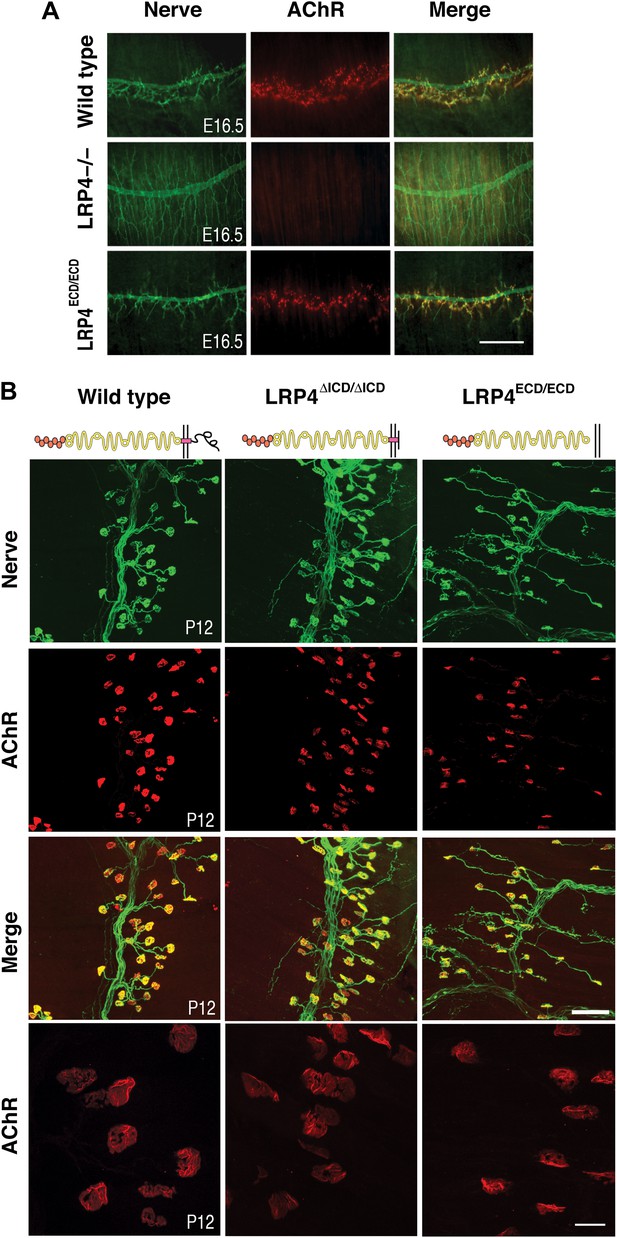
NMJ development in Lrp4 mutant mice.
(A) Embryonic diaphragm muscles (E16.5) from wild-type (top row), Lrp4–/– (middle row) and Lrp4ECD/ECD mice (bottom row) were double-labeled with anti-syntaxin antibodies (green) for decorating innervating fibers and with α-bungarotoxin (red) to detect AChRs. In both wild-type and Lrp4ECD/ECD muscles, AChRs are clustered along the central region juxtaposed to nerve terminals. By contrast, AChR clusters are completely absent and NMJs fail to form in Lrp4–/– muscles. (B) Triangularis sterni muscles at postnatal day 12 were stained for nerves in green (anti-NF150 and anti-Syt2 antibodies) and AChRs in red (α-bungarotoxin). In both wild-type (left column) and Lrp4ΔICD/ΔICD mice (middle column), neuromuscular synapses were distributed within a narrow band near the nerve trunk. By contrast, neuromuscular synapses in Lrp4ECD/ECD mice (right column) were distributed across a broader region of the muscle, as prolonged nerve branches extended from the nerve trunk. Furthermore, AChR clusters are markedly smaller in the Lrp4ECD/ECD muscle, compared with the wild-type or Lrp4ΔICD/ΔICD muscle (bottom row: high-power views of AChR clusters). Scale bars, A: 300 µm; B: 200 µm (top three rows) and 50 µm (bottom row).
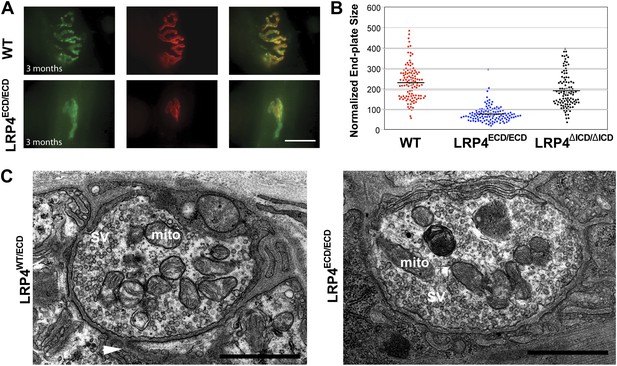
Reduced NMJ size in Lrp4ECD/ECD mice.
(A) Triangularis sterni (TS) muscles from wild type (WT) and 3-month-old Lrp4ECD/ECD mice double-labeled with anti-syntaxin antibodies (green, nerve) and α-bungarotoxin (red, AChRs). The NMJ in the Lrp4ECD/ECD muscle is markedly smaller than in the wild-type muscle. (B) Size comparison of neuromuscular synapse areas in TS muscles in 3-month-old wild-type, Lrp4ECD/ECD and Lrp4ΔICD/ΔICD mice. (C) Electron micrographs of NMJs from lumbrical muscles in 2-month old Lrp4WT/ECD and littermate Lrp4ECD/ECD mice. Nerve terminals containing numerous synaptic vesicles and mitochondria are embedded in the muscle surface and junctional folds are apparent in both genotypes. Scale bars, 1 μm. 55 nerve terminal profiles from 18 Lrp4ECD/ECD NMJs and 74 profiles from 23 control NMJs were analyzed. Representative images are shown. Arrow indicates junctional fold. SV: synaptic vesicle; mito: mitochondria.
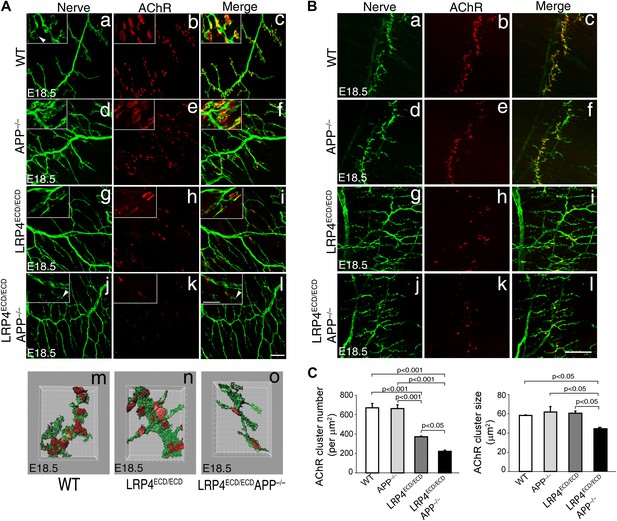
Impairment of pre-and post-synaptic development in Lrp4ECD/ECD;App–/– double mutant mice.
(A) (a–l) Wholemount staining of TS muscles (E18.5) double-labeled with anti-neurofilament antibodies and anti-Syt2 antibodies (nerve, a, d, g, j) and α-bungarotoxin (AChR, b, e, h, k). Merged images are shown in panels c, f, i and l. The inset in each panel shows a high-power view of the image. AChR clusters were markedly reduced both in number and size in Lrp4ECD/ECD;App−/− mice (j–l), compared to wild type (a–c), App−/− (d–f) or Lrp4ECD/ECD (g–i). Furthermore, numerous terminal sprouts (arrowheads in the inset of j and l) were seen in Lrp4ECD/ECD;App−/− mutant mice, whereas the nerve terminals in the wild type (arrowhead in a) juxtaposed with postsynaptic AChR clusters. m–o: 3D reconstruction of confocal images of the NMJs in wholemount diaphragm muscles in wild type (m), Lrp4ECD/ECD (n) and Lrp4ECD/ECD;App−/− (o) illustrate the reduced size of the NMJ in (o). Scale bars: a–l: 100 µm; inset: 20 µm. (B) Wholemount staining of diaphragm muscles (E18.5) double-labeled with anti-neurofilament antibodies and anti-Syt2 antibodies (nerve, a, d, g, j) and α-bungarotoxin (AChR, b, e, h, k). Low-power views of the left dorsal region of the diaphragm muscles. Scale bar, 400 µm. The number of AChR clusters was notably reduced in Lrp4ECD/ECD mice, compared to wild-type and App−/− mice. However both the size and number of AChR clusters were markedly reduced in Lrp4ECD/ECD;App−/− mice, compared to wild-type, App−/− and Lrp4ECD/ECD mice. (C) Quantitative analysis of AChR clusters size and numbers. The number of AChR clusters at the ventral regions of right hemi-diaphragm muscles (E18.5), normalized by muscle area (upper panel). Average size of AChR clusters (lower panel). The numbers of AChR clusters analyzed are: 144 wild type, 132 App−/−, 131 Lrp4ECD/ECD and 96 Lrp4ECD/ECD;App−/− (N = 3 mice for each genotype). Data are shown as average ± S.E.M. Pairwise multiple comparisons were carried out using Tukey’s test and the statistical differences determined by one-way analysis of variance (ANOVA).
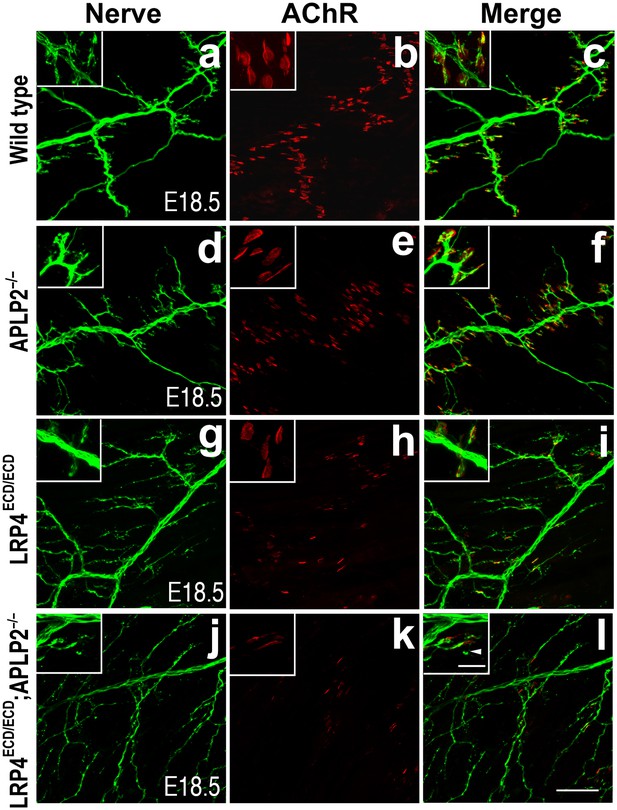
Impairment of pre-and post-synaptic development in Lrp4ECD/ECD;Aplp2–/– double mutant mice.
Wholemount staining of triangularis sterni muscles (E18.5) double-labeled with anti-neurofilament antibodies and anti-Syt2 antibodies (nerve, a, d, g, j) and α-bungarotoxin (AChR, b, e, h, k). Merged images are shown in panels c, f, i and l. The inset in each panel shows a high-power view of the image. The nerve terminals and AChR clusters were markedly reduced (both in number and size) in Lrp4ECD/ECD;Aplp2−/− mice (j–l), compared to wild type (a–c), Aplp2−/− (d–f) or Lrp4ECD/ECD (g–i). Arrowhead in the inset of l indicates a nerve terminal sprout. Scale bars: a–l: 100 µm; inset: 20 µm.
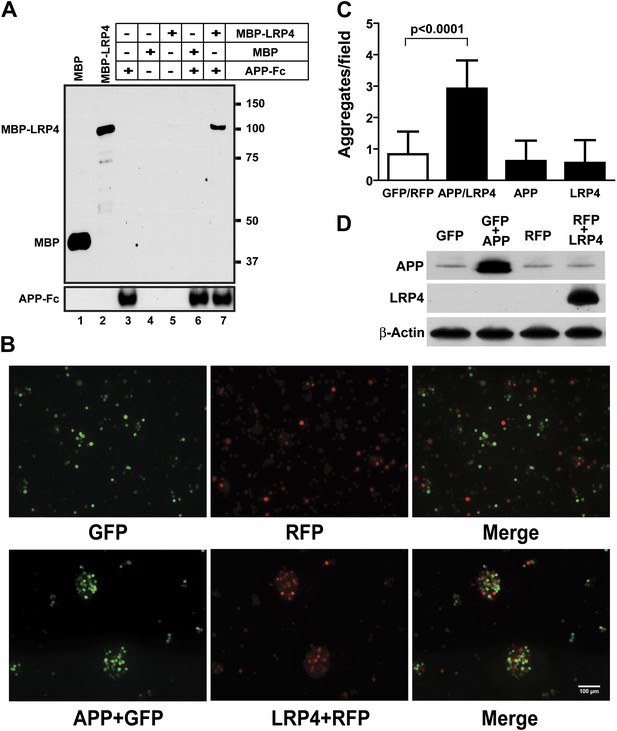
APP interacts with LRP4.
(A) APP-Fc-bead conjugates or blank beads were incubated with 100 ng purified MBP or MBP-LRP4LBD for 4 hr at 4°C prior to immunoprecipitation with anti-MBP followed by immunoblotting with anti-hFc antibody (lane 3–7). Lanes 1 and 2 show MBP (control protein) and MBP-LRP4LBD, respectively. (B) GFP- and RFP-expressing HEK293T or APP/GFP- and LRP4/RFP-expressing HEK293T cells were mixed in suspension and incubated for 30 min at 37°C before capturing cell aggregates under a fluorescent microscope. (C) The number of aggregates bigger than 3000 µm2 from twelve randomly selected images was determined. A significantly (p<0.0001) increased number of aggregates were present only when APP and LRP4 expressing cells were mixed. (D) APP and LRP4 protein levels in the transfected HEK293T cells.
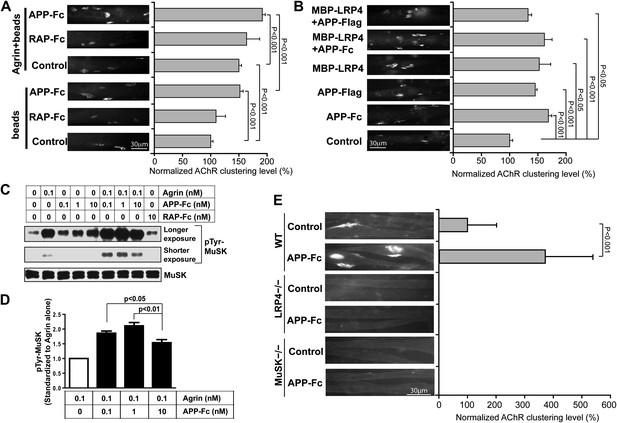
APP activates MuSK and promotes AChR clustering.
(A) C2C12 myotubes were incubated with the indicated beads preloaded with APP-Fc or RAP-Fc, respectively, in the absence or presence of 0.1 ng/ml agrin for 24 hr prior to labeling AChRs. The number of average AChR clusters per 200 µm myotube was counted from 50 randomly captured images and normalized to control levels. (B) Monomeric and divalent recombinant APP fusion proteins and MBP-LRP4, alone or in combination with APP fusion proteins, are equally effective in inducing AChR clustering. C2C12 myotubes were incubated with 1 µg/ml of soluble monovalent APP (APP-Flag), divalent APP (APP-Fc), or MBP-LRP4, alone or in combination with APP fusion proteins, for 24 hr prior to labeling AChRs. (C) Myotubes were incubated for 30 min at 37°C with the indicated relative molar concentrations of proteins prior to immunoblotting for total MuSK and tyrosine phosphorylated MuSK (pTyr-MuSK). 0.1 nM Agrin = 9 ng/ml, 0.1 nM APP-Fc = 13 ng/ml, 10 nM RAP-Fc = 720 ng/ml. (D) Relative amounts of pTyr-MuSK were quantified from three independent experiments. At all APP concentrations MuSK phosphorylation was significantly increased for agrin alone (0.1 nM APP, p=0.0003; 1 nM APP, p=0.0002; 10 nM APP, p=0.0031). (E) APP-induced AChR clustering requires LRP4 and MuSK. Primary myotubes cultured from wild-type (WT), Lrp4−/−, and Musk−/− mouse embryos (E18.5) were incubated in the absence (control) or presence of APP-Fc for 24 hr prior to labeling AChRs. Statistical analysis by Student’s t-test. Scale bars, 30 µm.
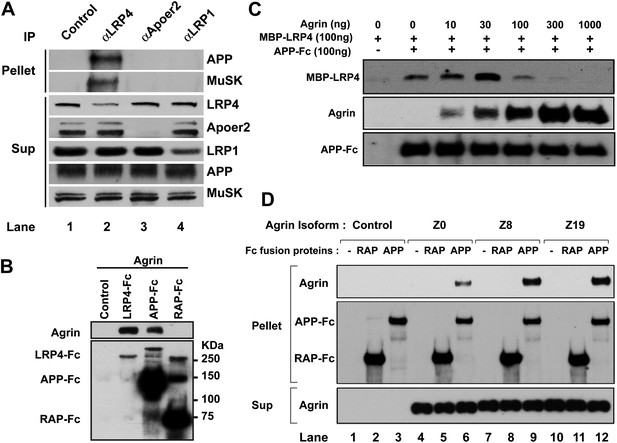
Interactions between agrin, APP and LRP4.
(A) APP, LRP4 and MuSK form a stable complex in muscle in vivo. Proteins were extracted from wild-type embryo (E14.5) muscles and incubated with polyclonal anti-LRP4, anti-Apoer2, or anti-LRP1 antibodies overnight at 4°C followed by adsorption to Protein A Dynabeads. Supernatants were efficiently immunodepleted of LRP4, Apoer2 and LRP1 with the respective antibodies. APP and MuSK coprecipitate with LRP4, but not with the other LRP members Apoer2 and LRP1, indicating the presence of a physiological complex consisting of LRP4, APP and MuSK in muscle in vivo. (B) Control (non-transfected), LRP4-Fc, APP-Fc, or RAP-Fc containing HEK293T supernatants were adsorbed with Protein A sepharose beads. Conjugated beads were incubated with 1 µg neural agrin in a final volume of 1 ml for 4 hr at 4°C, followed by immunoblotting for bound Fc proteins and agrin. (C) APP-Fc-bead conjugates or blank beads were incubated with purified 100 ng of MBP-LRP4LBD and 0–1000 ng of neural agrin for 4 hr at 4°C. The precipitate was analyzed by immunoblotting with anti-MBP, anti-agrin, and anti-Fc antibodies to detect cooperative protein interactions among LRP4, APP, and agrin. (D) Control (lanes 1–3), agrin Z0 (lanes 4–6), agrin Z8 (lanes 7–9), or agrin Z19 (lanes 10–12) containing culture supernatants were incubated overnight at 4°C in the absence (lanes 1, 4, 7, 10) or presence of RAP-Fc (lanes 2, 5, 8, 11) or APP-Fc (lanes 3, 6, 9, 12) prior to adsorption to Protein A coupled Dynabeads and magnetic isolation. Bound agrin isoforms and Fc proteins were determined by immunoblotting with anti-Myc and anti-Fc antibodies.
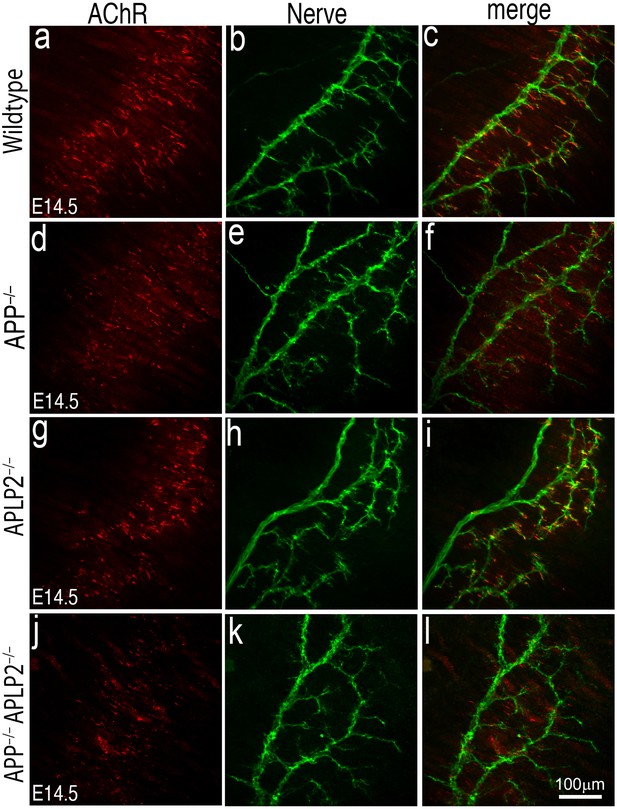
AChR prepatterning in the absence of APP and APLP2.
Wholemounts of embryonic diaphragm muscles (E14.5) from wild type (a–c), App–/– (d–f), Aplp2–/– (g–i) and App–/–;Aplp2–/– (j–l) embryos were double-labeled with α-bungarotoxin (red) and anti-syntaxin antibodies (green). In wild-type and all mutant embryos, AChR clusters were confined to the central regions of diaphragm muscles indicating AChR prepatterning is independent of APP and APLP2. However, individual AChR clusters appear less robust in App–/–;Aplp2–/– muscle compared with those in the wild-type, App–/– or Aplp2–/– muscles. Scale bar: 100 μm.
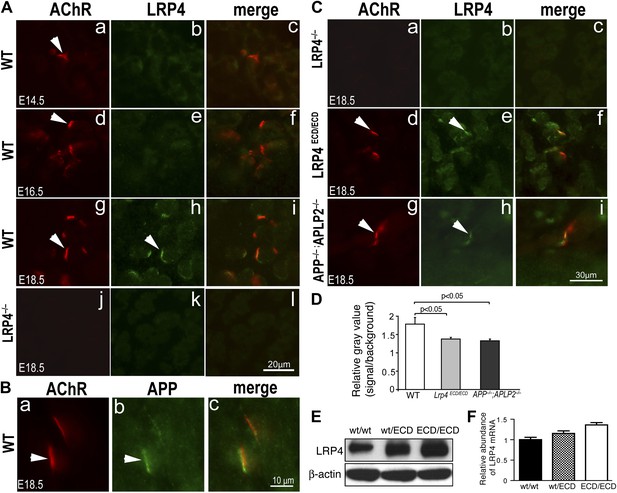
LRP4 localization in Lrp4ECD/ECD and App–/–;Aplp2–/– mutant mice.
(A) Cross sections of hindlimb muscle from E14.5 (a–c), E16.5 (d–f) and E18.5 (g–i) wild type (WT) and E18.5 Lrp4–/– (j–l) mice were double-labeled with anti-LRP4 antibodies (green) and Texas-Red conjugated α-bungarotoxin (red), which marks the site of the NMJ (arrowhead in a, d, g). LRP4 staining appears at low level in E14.5 WT and E16.5 WT muscle (compared with Lrp4–/– muscle), and become highly concentrated at the NMJ in E18.5 WT muscle (arrowhead in h). Scale bar in A: 20 µm. (B) Localization of APP at the NMJ. Wholemount diaphragm muscle from E18.5 WT mice was double-labeled with anti-APP antibodies (b) and Texas-Red conjugated α-bungarotoxin (a). APP (arrowhead in b) is localized at the NMJ (arrowhead in a), as shown in the merged image in c. Scale bar in B: 10 µm. (C) Cross sections of hindlimb muscles from Lrp4–/– (a–c), Lrp4ECD/ECD (d–f) and App–/–;Aplp2–/– (g–i) mice (E18.5) were double-labeled with anti-LRP4 antibodies (green) and α-bungarotoxin (red). LRP4 was detected at the NMJ in Lrp4ECD/ECD (arrowhead in e) and App–/–;Aplp2–/– (arrowhead in h), but not in Lrp4–/– muscle (b). LRP4 expression appeared diffused in Lrp4ECD/ECD and in App–/–;Aplp2–/– muscles compared with age-matched WT muscle (see A). Scale bar in C: 30 μm. (D) Quantification of relative fluorescence intensity for LRP4 immunostaining on hindlimb myofibers from WT, Lrp4ECD/ECD and App–/–;Aplp2–/– mice (E18.5). Gray value of LRP4 staining at the NMJ (defined as signal) and within the sarcoplasm (defined as background) of the same myofiber was separately measured using ImageJ. The ratio of signal to background was then calculated for each individual myofiber. The bar graph shows significant (p<0.05) decreases in LRP4 staining in Lrp4ECD/ECD (1.38 ± 0.05, n = 23 myofibers) and App−/−;Aplp2−/− (1.33 ± 0.05, n = 22 myofibers), compared with WT muscle (1.78 ± 0.18, n = 19 myofibers). (E) Anti-LRP4 and beta-actin (loading control) immunoblot shows increased level of LRP4 protein expression in Lrp4ECD/ECD and Lrp4WT/ECD muscle, compared with WT muscle (from the hindlimb muscles of 10-week-old mice). (F) Lrp4 mRNA expression in hindlimb muscles (10-week-old mice) was determined by quantitative PCR. Levels of Lrp4 mRNA were normalized to cyclophilin mRNA levels. Results are shown as average ± SD of triplicates.
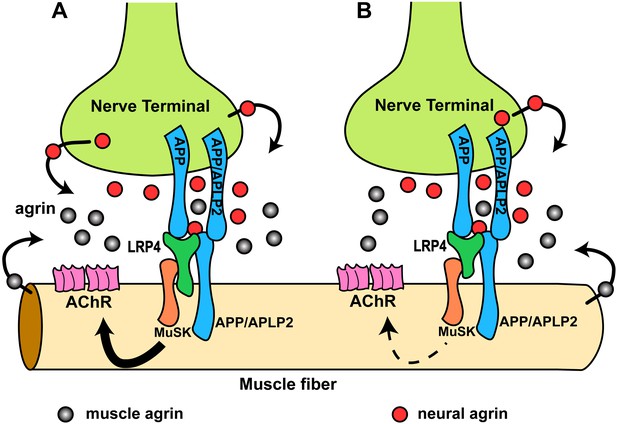
Hypothetical model of the interactions of APP/APLP2, LRP4, agrin and MuSK during NMJ formation.
(A) Our genetic, biochemical and functional results suggest that APP interacts with LRP4 and agrin to regulate NMJ formation. APP–LRP4 interaction in the absence of agrin may be able to activate MuSK signaling (Figure 7C) at low levels to promote AChR clustering on the muscle fiber membrane, raising the possibility that nerve and muscle-derived APP and APLP2 homo- or heterodimerization may be able to cooperatively promote AChR clustering in the absence of agrin, which provides a potential mechanism for the earlier observation that initial AChR clustering is independent of agrin (Lin et al., 2001; Yang et al., 2001). Neural agrin (red) enhances the APP–LRP4 interaction and strongly activates MuSK (Figures 7C and 8C), which is consistent with the role of agrin in the stabilization and maintenance of NMJs. Cooperative binding of APP and agrin to LRP4 (Figure 8C) would further strengthen LRP4–APP interaction on muscles, promoting synaptic differentiation and postnatal maintenance of NMJs (solid line with arrow in muscle fiber cytoplasm). By contrast, muscle agrin (gray), which cannot activate MuSK, would compete with decreasing concentrations of diffusible neural agrin at increasing distance from the NMJ and thus keep AChR clustering outside the nerve contact site suppressed. (B) Loss of the LRP4 membrane anchor results in a secreted ectodomain that remains partially functional through bivalent interactions with MuSK (Zhang et al., 2008; Kim et al., 2008b) and APP, resulting in reduced signaling (dashed line with arrow) and thus a hypomorphic NMJ developmental defect compatible with embryonic NMJ formation, but impaired postnatal maturation and maintenance of the NMJ.
Tables
Postnatal survival of Lrp4ECD/ECD;App−/− double mutant mice
| Genotype | Expected | Observed | Chi-square test |
|---|---|---|---|
| Lrp4+/ECD;App+/− | 63 | 74 | n.s. |
| Lrp4+/ECD;App−/− | 44 | 45 | n.s. |
| Lrp4ECD/ECD;App+/− | 49 | 49 | n.s. |
| Lrp4ECD/ECD;App−/− | 38 | 15 | * |
-
Note: 41% (20/49) of Lrp4ECD/ECD;App+/− died within five months of age; 13% (6/45) of Lrp4+/ECD;App−/− mice died within five months of age.
-
n.s., not significant.
-
*
Significant at p<0.01. Survival was scored at weaning (P25). The ‘Expected’ column states the number of offspring of the indicated genotype according to the rules of Mendelian inheritance. The ‘Observed’ column states the number of offspring found to have the indicated genotype. Chi-square analysis of these data indicates whether the observed number significantly deviates from the expected number or not.
Survival analysis of Lrp4ECD/ECD;Aplp2−/− double mutant pups
| Genotype | Expected | Observed | Chi-square test |
|---|---|---|---|
| Lrp4+/ECD;Aplp2+/− | 68 | 82 | n.s. |
| Lrp4+/ECD;Aplp2−/− | 69 | 75 | n.s. |
| Lrp4ECD/ECD;Aplp2+/− | 58 | 51 | n.s. |
| Lrp4ECD/ECD;Aplp2−/− | 54 | 17 | * |
-
n.s., not significant.
-
*
Significant at p<0.01.
-
Pups were genotyped at weaning (∼25 days of age) to determine postnatal survival rates.





