Hypothemycin, a fungal natural product, identifies therapeutic targets in Trypanosoma brucei
Figures
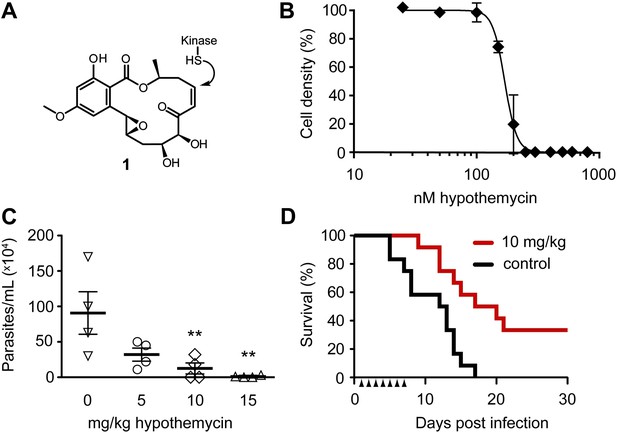
Potent trypanocidal activity of hypothemycin.
(A) Hypothemycin (1) inhibits CDXG kinases via conjugate addition to the cis-enone. (B) Bloodstream form T. brucei were treated with hypothemycin and cell density was measured after 24 hr (mean ± SD, n = 3). (C) Parasitemia in T. brucei infected mice. Mice received once daily intraperitoneal injections of hypothemycin and parasitemia was measured 5 days post infection (mean ± SD, n = 4, ** denotes p<0.05). (D) Kaplan-Meier analysis of T. brucei infected mice. Hypothemycin or vehicle was administered by intraperitoneal injection once daily for 7 days post infection (arrowheads, n = 12). Data were accumulated from three studies, p<0.01.
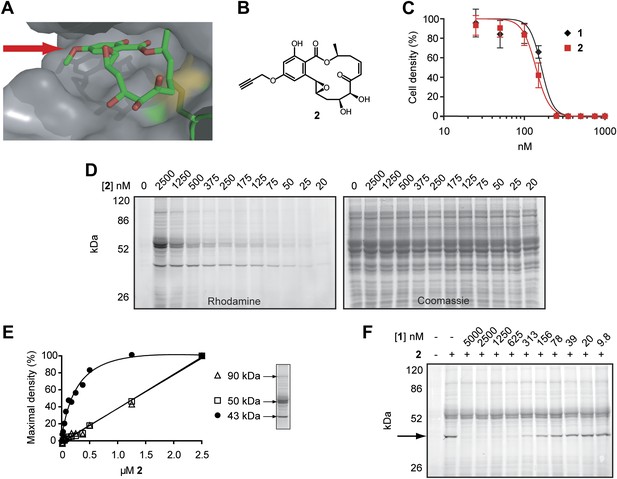
Design and validation of a hypothemycin-based affinity probe.
(A) Crystal structure of hypothemycin (1) bound to ERK2 (PDB: 3C9W), indicating the solvent-exposed C4 methyl ether. (B) Structure of probe 2. (C) Effect of 2 on proliferation of cultured BSF T. brucei (mean ± SD, n = 3). (D) T. brucei whole-cell lysates were treated with 2 for 30 min. Labeled proteins were visualized after click conjugation to rhodamine-azide and separation by SDS-PAGE. (E) Fluorescence quantification of bands at 90, 50, and 43 kDa demonstrating saturation of the 43 kDa band, but not the 90 or 50 kDa bands. (F) Lysates were treated with the indicated concentrations 1 for 30 min, followed by 500 nM 2 for 30 min. Labeled proteins were visualized as above. The saturable 43 kDa protein is indicated by the arrow.
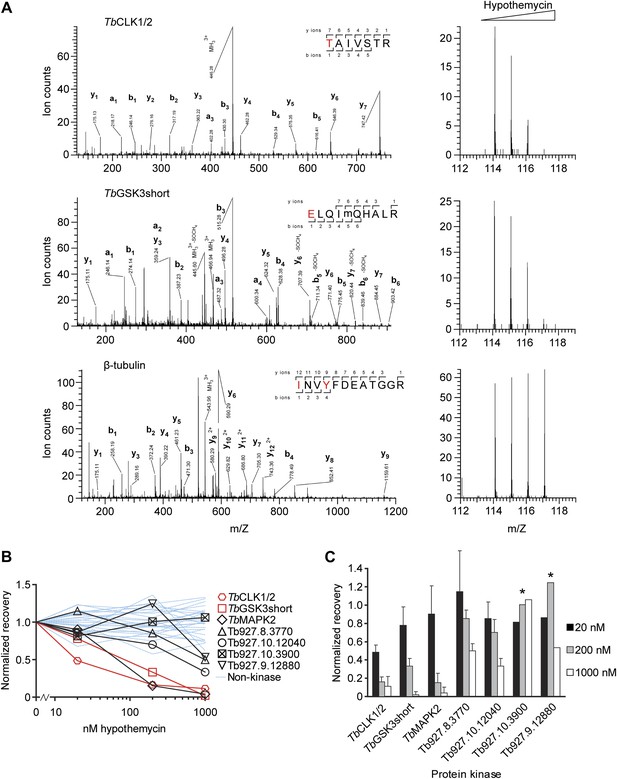
Quantification of hypothemycin binding to CDXG kinases.
(A) MS/MS fragmentation spectra of peptides from TbCLK1/2, TbGSK3short, and β-tubulin after labeling with 2, affinity purification, and trypsinization. Red text: residues bearing an iTRAQ tag; ‘m’: oxidized Met. iTRAQ reporter ions for each peptide are shown to the right (m/Z 114, 115, 116, and 117 correspond to lysate samples pretreated with 0, 20, 200, or 1000 nM hypothemycin, respectively). (B) Normalized recovery values, based on iTRAQ quantification, for all identified protein kinases and 25 non-kinases (blue lines) with ≥3 unique peptides. (C) Recovery of protein kinases as a function of hypothemycin pretreatment. Values for each hypothemycin pretreatment condition are derived from mean iTRAQ reporter ion counts normalized to vehicle across all peptides identified (± SD). Asterisks indicate values derived from a single spectrum.
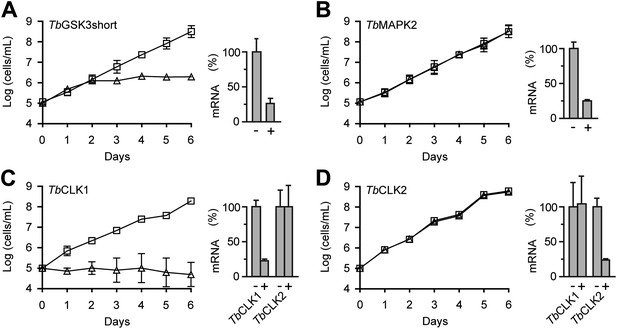
RNAi analysis of CDXG kinases.
(A–D) BSF T. brucei were stably transfected with the indicated RNAi constructs and induced with tetracycline (triangles) or left uninduced (squares) starting on day 0. Cell density was measured every 24 hr and cumulative cell growth was plotted on a log scale (mean ± SD, n = 3). mRNA levels were measured by quantitative RT-PCR (bar graphs) in the absence (–) and presence (+) of tetracycline.
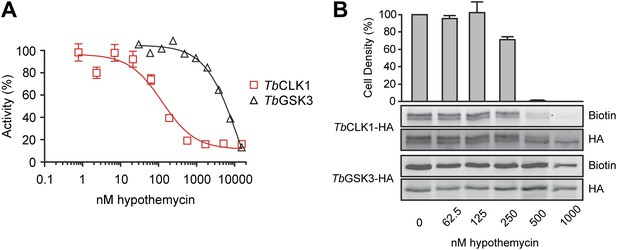
Preferential inhibition of TbCLK1 by hypothemycin.
(A) In vitro assays with recombinant TbGSK3short and TbCLK1. Kinases were incubated with hypothemycin and 100 μM ATP for 30 min before initiating reactions with substrate and γ32P-ATP. Substrate phosphorylation was quantified and normalized to DMSO control (mean ± SD, n = 3). (B) T. brucei expressing HA-tagged TbGSK3short and TbCLK1 from their endogenous loci were incubated with hypothemycin for 5 hr, and then were either harvested or diluted 1:30 into drug-free media and counted after 24 hr (bar graph, mean ± SD, n = 3). Harvested cells were lysed, labeled with 2, and submitted to click conjugation with biotin-azide. HA-tagged proteins were immunoprecipitated, resolved by SDS-PAGE, and analyzed by Western blotting for biotin and HA.
Additional files
-
Supplementary file 1
(A) Mass Spectrometry Results. (B) Summary of RNAi results. (C) Primers used in this study.
- https://doi.org/10.7554/eLife.00712.008





