ATR/Mec1 prevents lethal meiotic recombination initiation on partially replicated chromosomes in budding yeast
Figures
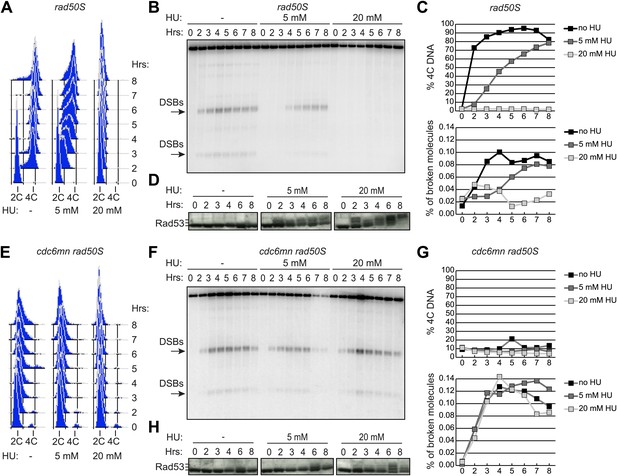
Ongoing DNA replication delays meiotic DSB formation.
rad50S (H156, A–D) or cdc6-mn rad50S (H155, E–H) cells were induced to enter meiosis in 0, 5 or 20 mM HU and analyzed at the indicated time points. (A and E) FACS analysis of total DNA content. (B and F) Southern blot analysis of DSB formation at the yCR048W DSB hotspot. Arrows indicate the major DSB bands quantified in (C and G). (C and G) Quantification of 4C DNA content from FACS is shown in the upper panel. The measurement of DSBs from Southern blot is plotted in the lower panel. (D and H) Western blot analysis of Rad53 protein mobility is shown as a measurement of phosphorylation and activation. Slower migrating bands correspond to phosphorylated Rad53. See also Figure 1—figure supplement 1.
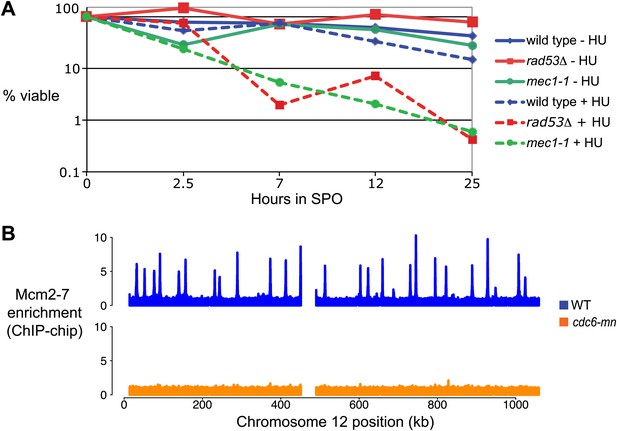
Replication and checkpoint requirements during meiS.
(A) The viability of sml1Δ cells (H3554), mec1-1 sml1Δ (H2560) and rad53Δ sml1Δ (H2591) cells is plotted with respect to time in SPO. The number of viable colonies was normalized to the number at 0 hr for each culture. (B) Mcm2-7 genome-wide location (ChIP-chip) analysis is presented for wild-type (H2544, Blitzblau et al., 2012) and cdc6-mn (H154) cells as indicated.
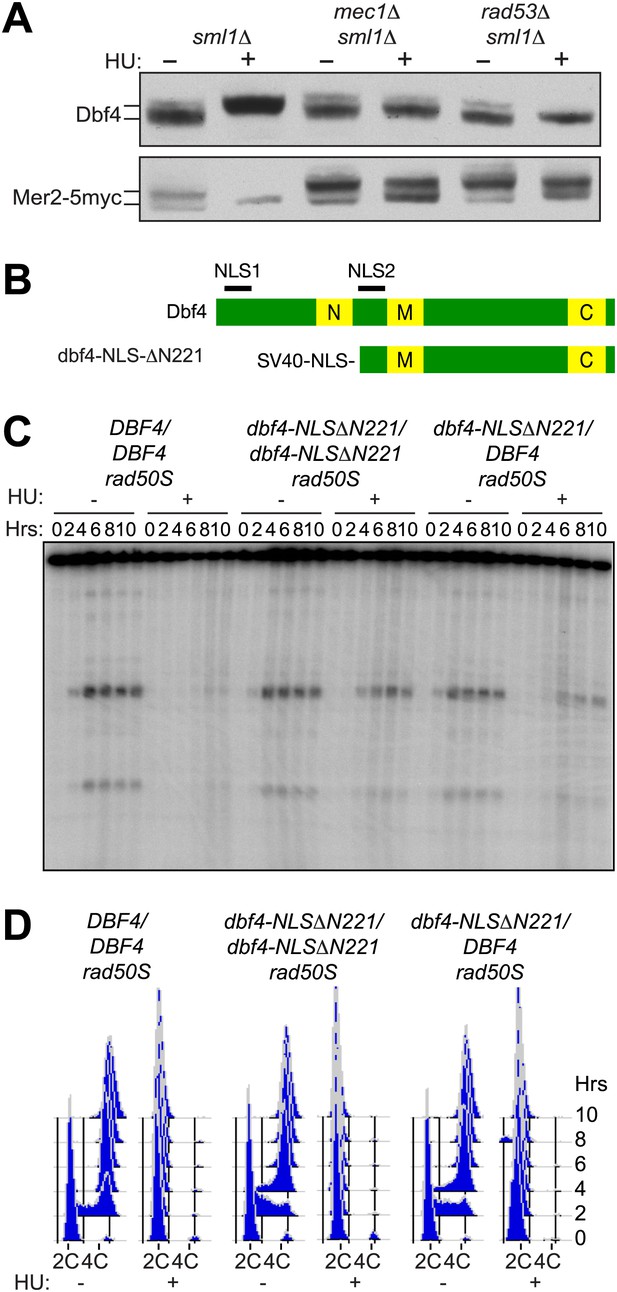
The pre-meiotic replication checkpoint inhibits DDK kinase activity.
(A) Western blot analysis of Dbf4 (top panel) and Mer2-5myc (bottom panel) in sml1Δ (H5157), mec1Δ sml1Δ (H5220) and rad53Δ sml1Δ (H5127) cells. The sml1Δ mutation was used to maintain viability of mec1Δ and rad53Δ mutants. For Mer2-5myc blotting, only 20% (wild-type) or 50% (mec1Δ and rad53Δ) of total protein was loaded for HU-treated samples as high accumulation of the Mer2 protein obscured the analysis of mobility shifts. (B) Schematic of wild-type and mutant Dbf4 proteins analyzed in this study. N, M, and C refer to the N-terminal, middle and C-terminal conserved domains. (C) Southern blot analysis of DSB formation at the yCR048W DSB hotspot in rad50S cells for DBF4/DBF4 (H6097), dbf4-NLS-ΔN221/dbf4-NLS-ΔN221 (H6146) and dbf4-NLS-ΔN221/DBF4 (H7335) cells. (D) FACS analysis of total DNA content of the strains in (C). See also Figure 2—figure supplement 1.
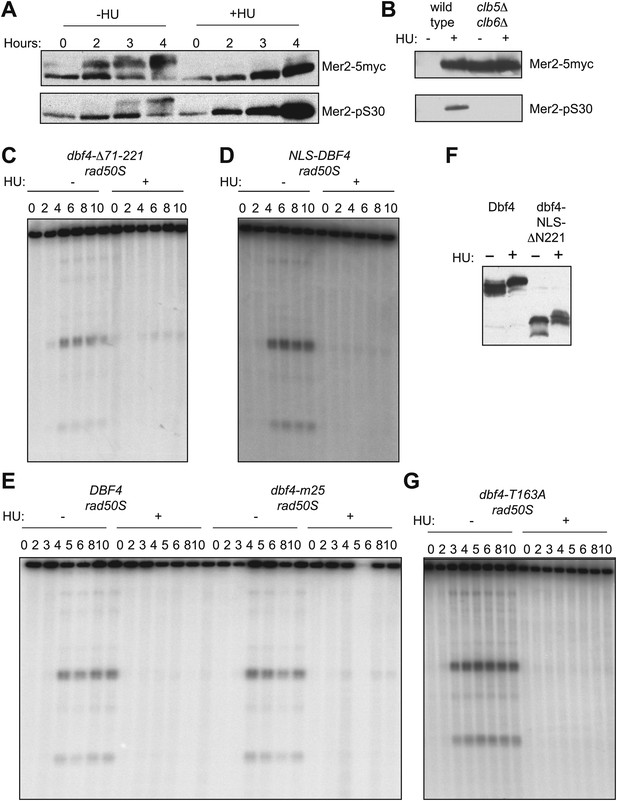
Regulation of DDK prevents DSBs in HU-treated cells.
(A) Western blot analysis of Mer2-5myc in an ndt80Δ spo11-Y135F-HA strain (H5079) probed with anti-myc (top panel) for total protein and anti-Mer2 phospho-S30 antibody (bottom panel) in the absence or presence of HU. (B) Specificity of the anti-Mer2 phospho-S30 antibody was confirmed by comparing the signals on proteins from wild-type (H4695) and clb5Δ clb6Δ (H5076) cells after 5 hr in SPO medium by Western blotting using the same conditions as (A). These cells are NDT80 (i.e., not blocked in prophase), so Mer2 is largely degraded in wild-type cells without HU treatment. (C) Southern blot analysis of DSBs at the yCR048W hotspot in dbf4-Δ71-221 rad50S cells (H6296). (D) Southern blot analysis of DSBs at the yCR048W hotspot in NLS-DBF4 rad50S cells (H7309). (E) Southern blot analysis of DSBs at the yCR048W hotspot in rad50S (H4226) and dbf4-m25 rad50S (H5603) cells. (F) Western blot analysis of Dbf4 (H6097) and dbf4-ΔN221 (H6146) proteins in the absence or presence of HU. (G) Southern blot analysis of DSBs at the yCR048W hotspot in dbf4-T163A rad50S (H4883) cells.
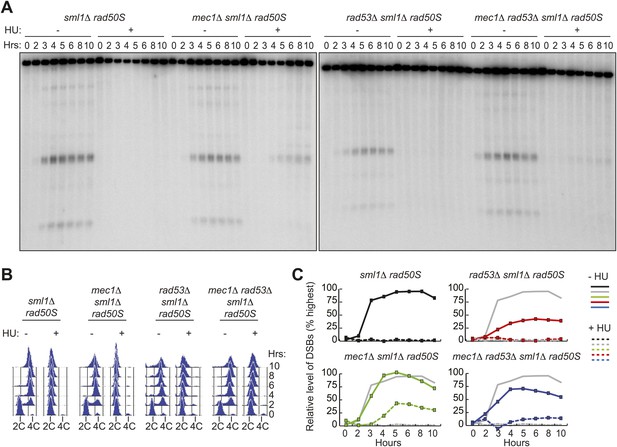
Removal of MEC1, but not RAD53, allows DSB formation in HU-treated cells.
(A) Southern blot analysis of DSB formation at the yCR048W DSB hotspot in sml1Δ rad50S (H4898), mec1Δ sml1Δ rad50S (H4935) and rad53Δ sml1Δ rad50S (H4969) and mec1Δ rad53Δ sml1Δ rad50S cells (H4932) in the absence or presence of HU. (B) FACS analysis of total DNA content of the strains in (A). (C) Quantification of the relative DSB levels from the Southern blot in (A) in the absence (solid lines) or presence (dashed lines) of HU. DSBs levels were measured as in Figure 1 and normalized to the maximum measurement in the sml1Δ rad50S (H4898) ‘wild-type’ strain (shown as grey lines for comparison). See also Figure 3—figure supplement 1.
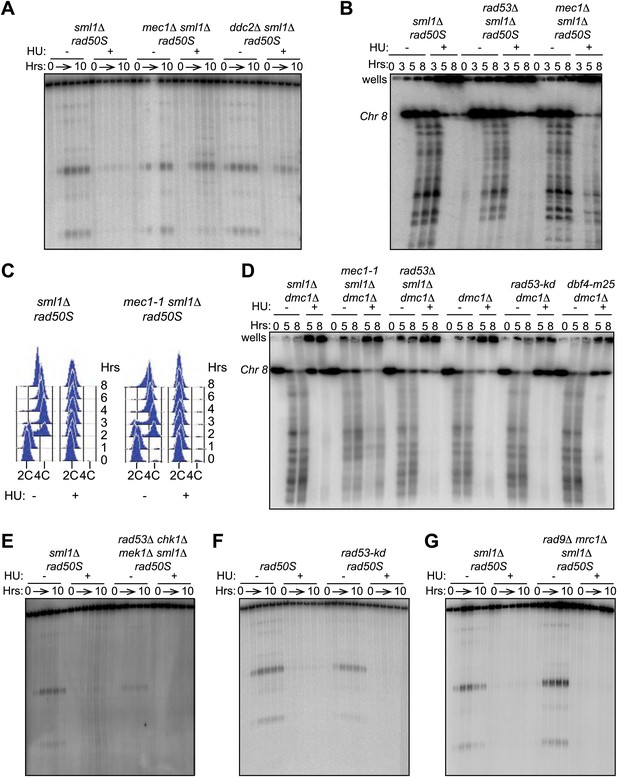
Removing Mec1, but not Rad53, allows DSBs in HU-treated cells.
(A) Southern blot analysis of DSBs at the yCR048W hotspot in sml1Δ rad50S (H4898), mec1Δ sml1Δ rad50S (H4935) and ddc2Δ sml1Δ rad50S (H6002) strains. (B) CHEF gel electrophoresis and Southern blot analysis of DSB formation on Chromosome 8 in sml1Δ rad50S (H4898), rad53Δ sml1Δ rad50S (H4969), and mec1Δ sml1Δ rad50S cells (H4935). The probe used for Southern blotting in SGD coordinates was: Chromosome VIII: 23,771-25,410. (C) FACS analysis of total DNA content in sml1Δ rad50S (H4898) and mec1-1 sml1Δ rad50S (H4557) strains. (D) CHEF gel electrophoresis and Southern blot analysis of DSB formation on Chromosome 8 in sml1Δ dmc1Δ (H4618), mec1-1 sml1Δ dmc1Δ (H4557) rad53Δ sml1Δ dmc1Δ (H6813), dmc1Δ cells (H118), rad53-kd dmc1Δ (H6815) and dbf4-m25 dmc1Δ (H6814) cells. The probe used for Southern blotting in SGD coordinates was: Chromosome VIII: 23,771-25,410. (E) Southern blot analysis of DSBs at the yCR048W hotspot in sml1Δ rad50S (H4898) and rad53Δ chk1Δ mek1Δ sml1Δ rad50S (H5241) cells. (F) Southern blot analysis of DSBs at the yCR048W hotspot in rad50S (H4226) and rad53-kd rad50S (H5884) cells. (G) Southern blot analysis of DSBs at the yCR048W hotspot in sml1Δ rad50S (H4898) and mrc1Δ rad9Δ sml1Δ rad50S (H5776) cells.
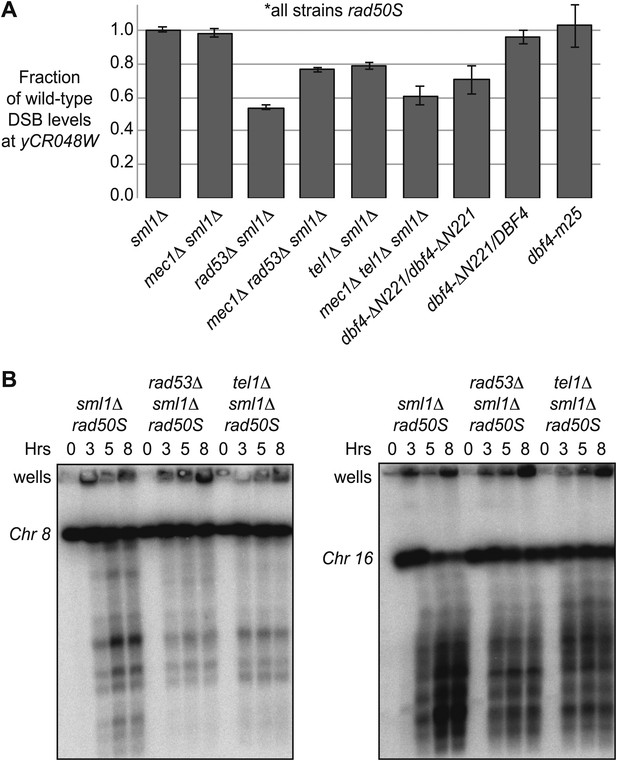
A Tel1-dependent feedback mechanism increases DSB levels in rad50S cells.
(A) The maximum levels of DSBs in untreated rad50S strains containing the indicated mutations were measured from the Southern blots shown in Figure 2, Figure 3, Figure 2—figure supplement 1, Figure 3—figure supplement 1, and Figure 4—figure supplement 1. The amount of broken DNA was calculated as in Figure 1 and all values were normalized to the wild-type strain from the same experiment. (B) CHEF gel electrophoresis and Southern blotting was conducted to assess DSB levels on whole chromosomes in sml1Δ rad50S (H4898), rad53Δ sml1Δ rad50S (H4969), and tel1Δ sml1Δ rad50S (H4849) cells. Chromosomes 8 (left panel) and 16 (right panel) were resolved on separate gels, blotted and visualized with the following probes in SGD coordinates: Chromosome VIII: 23,771-25,410 and Chromosome XVI: 20,281-21,012. See also Figure 4—figure supplement 1.
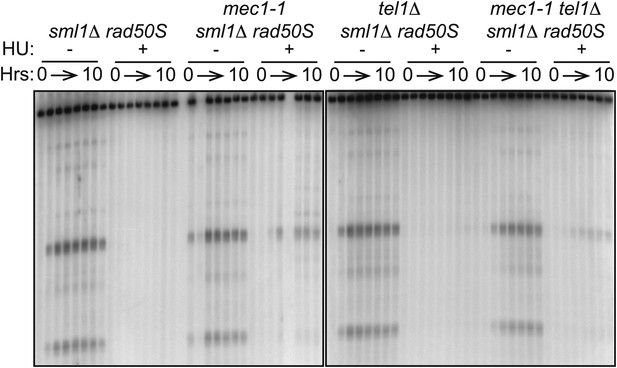
TEL1 is required for wild-type DSB levels in rad50S cells.
Southern blot analysis of DSBs at the yCR048W hotspot in sml1Δ rad50S (H4850), mec1-1 sml1Δ rad50S (H4851), tel1Δ sml1Δ rad50S (H4849) and mec1-1 tel1Δ sml1Δ rad50S (H4853) cells.
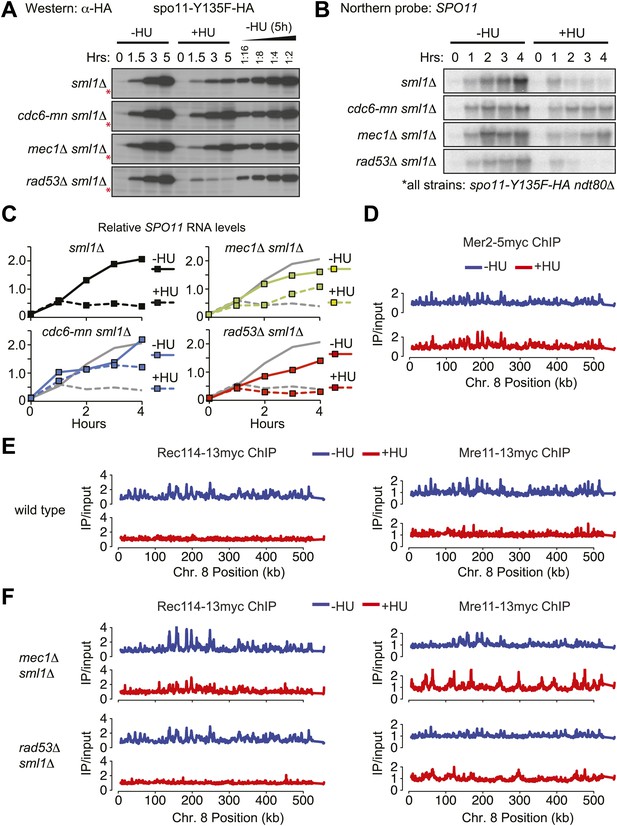
The replication checkpoint regulates DSB factor levels and DNA loading.
(A–C) Analysis of spo11-Y135F-HA in sml1Δ ndt80Δ (H5233), cdc6-mn sml1Δ ndt80Δ (H7447), mec1-1 sml1Δ ndt80Δ (H5227), rad53Δ sml1Δ ndt80Δ (H5230) strains in the presence or absence of HU. (A) Western blot analysis of spo11-Y135F-HA levels. A twofold dilution series of the 5h (–HU) time point was used to estimate changes in protein levels in the presence of HU. The cross-reacting band marked with red asterisks serves as a loading control. (B) Northern blot analysis of spo11-Y135F-HA RNA. (C) Quantification of the Northern blots in (B). Northern blots were reprobed for UBC6 (Teste et al., 2009) for normalization. RNA levels of the spo11-Y135F-HA sml1Δ ndt80Δ ‘wild-type’ strain are shown as grey lines in all panels for comparison. (D) Binding profiles for Mer2-13myc (H4585) from genome-wide location analysis (ChIP-chip) along Chromosome 8 in the absence (blue lines) or presence (red lines) of HU. (E) As in (D), binding profiles from ChIP-chip analysis for Rec114-13myc (H4890) and Mre11-13myc (H5547) in wild-type cells in the absence (blue lines, Vader et al., 2011) or presence (red lines) of HU. (F) ChIP-chip binding profiles of and Rec114-13myc in mec1Δ sml1Δ (H7305) and rad53Δ sml1Δ (H7302) cells and for Mre11-13myc in mec1Δ sml1Δ (H7323) and rad53Δ sml1Δ (H7320) cells in the absence (blue lines) or presence (red lines) of HU. See also Figure 5—figure supplement 1.
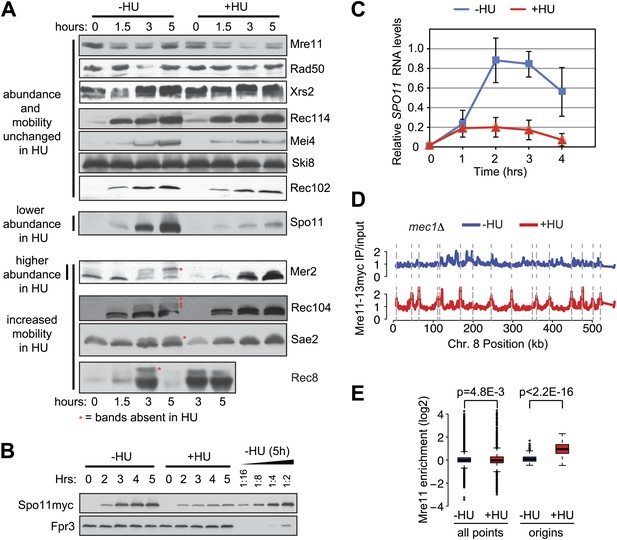
The meiotic replication checkpoint regulates DSB factor abundance, phosphorylation and DNA binding.
(A) Western blot analysis in the absence or presence of HU in ndt80Δ spo11-Y135F strains for: Mre11-13myc (H5085), Rad50-6HA (H7312), Xrs2-13myc (H5098), Rec114-13myc (H5092), Mei4-13myc (H5095), Ski8-13myc (H5154), Rec102-13myc (H5249), spo11-Y135F-HA (H5082), Mer2-5myc (H5079), Rec104-13myc (H5088), and Sae2-13myc (H5082). Rec8-3HA (H4695) cells are not ndt80Δ spo11Y135F. Red asterisks indicate the presence of bands that are absent in the HU-treated cells. (B) Western blot analysis in the absence or presence of HU for Spo11-18myc (H2087) in otherwise wild-type cells, therefore not ndt80Δ spo11Y135F. A twofold dilution series of the 5h (–HU) time point was used to estimate changes in protein levels in the presence of HU. Fpr3 serves as loading control. (C) Time course of SPO11 transcripts accumulation in Spo11-18myc cells (H2087) in the presence or absence of HU as determined by Northern blotting. SPO11 signals were normalized to FPR4 transcript levels. Shown are the mean and standard deviations of four independent Northern assays. (D) ChIP-chip binding profiles for Mre11-13myc in mec1Δ sml1Δ (H7323) cells in the absence (blue) or presence (red) of HU are shown for Chromosome 8. The sites of pre-meiotic replication origins are indicated by vertical dashed grey lines. (E) The distribution of values of either all points or replication origins (as indicated) were plotted for the Mre11-13myc binding profiles in mec1Δ sml1Δ (H7323) cells in the absence (blue) or presence (red) of HU. The p values (Student's t test) of the differences between the distributions are shown above the plot.
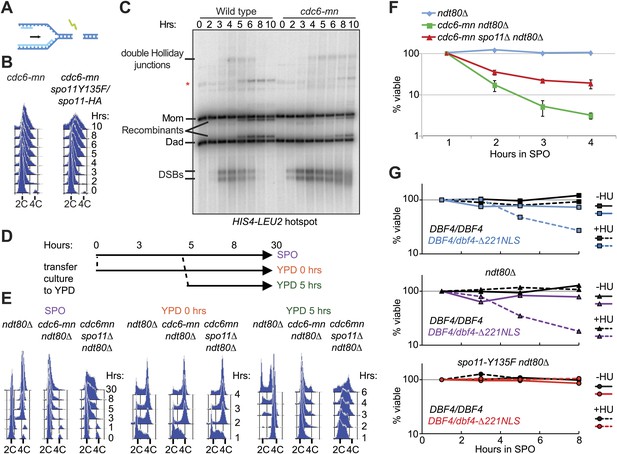
DSBs on replicating chromosomes are lethal.
(A) Schematic of a DSB that occurs ahead of a DNA replication fork. (B) FACS analysis of total DNA content in cdc6-mn (H2655) and cdc6mn spo11-HA3-His6/spo11-Y135F (H3598) cells as they progress through meiosis. (C) Southern blot analysis of DSB formation and repair at the HIS4-LEU2 hotspot in wild-type (H2636) and cdc6-mn (H2655) cells. Cells were treated with psoralen and DNA was crosslinked with UV light to preserve recombination intermediates. The relative positions of parental bands, repair intermediates and recombinants are marked. The red asterisk marks the position of an alternative recombination product. (D) Schematic of the experiment shown in (E and F). Cells from ndt80Δ (H385), cdc6-mn ndt80Δ (H386) and cdc6-mn spo11Δ ndt80Δ (H3682) pre-sporulation cultures were split into YPD or SPO to induce mitosis or meiosis, respectively. After 5 hr, half of the SPO culture was returned to growth in YPD. (E) FACS analysis of total DNA content was performed for the experiment described in (D). (F) Viability of cells was measured for the strains described in (D) at the indicated time points during meiotic induction. The number of colonies at each time point was normalized to the 1-hr time point for each culture. (G) Viability of wild-type (H7099), dbf4-NLS-ΔN221/DBF4 (7401), ndt80Δ (H7494), dbf4-NLS-ΔN221/DBF4 ndt80Δ (H7493), ndt80Δ spo11-Y135F-HA (H7468) and dbf4-NLS-ΔN221/DBF4 ndt80Δ spo11-Y135F-HA (H7469) strains induced to enter meiosis in the presence or absence of HU and transferred onto YPD medium at the indicated time points. Each point is the average of two or three independent experiments. Viabilities were normalized to the 1-hr time point for each culture. See also Figure 6—figure supplement 1.
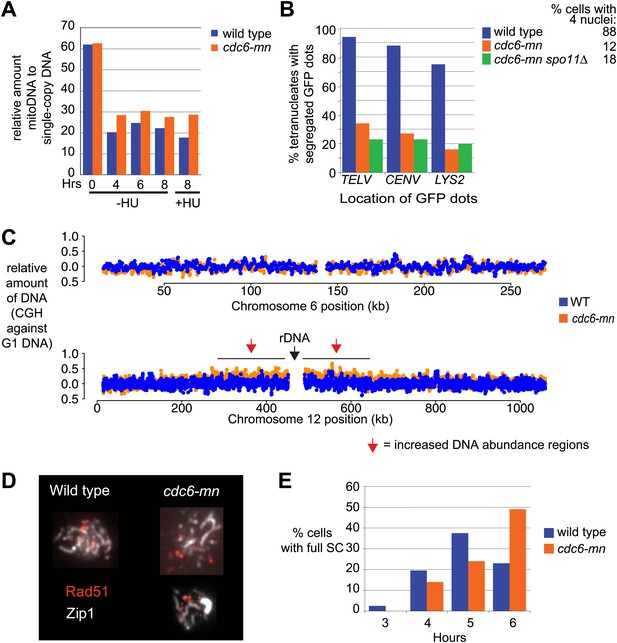
Characterization of DNA replication and SC formation in cdc6-mn cells.
(A) Relative copy number of mitochondrial DNA to single-copy chromosomal DNA in wild-type (H2775) and cdc6-mn (H2776) cells in the presence or absence of HU calculated by Southern blot analysis of HaeIII-digested genomic DNA probed for COX2 for the mitochondrial DNA and CEN15 for single-copy DNA. (B) Cells containing a single GFP-marked chromosome using TetR-GFP and a TetO array integrated on one homolog at TELV, CENV or LYS2, respectively, for wild-type (H3758, H3755, H3805, blue bars), cdc6-mn (H3756, H3753, H3803, orange bars) and cdc6-mn spo11Δ (H3757, H3754, H3804, green bars) were analyzed after 24-hour incubation in SPO. The number of GFP dots in each tetranucleate cell was counted as a measure of the ability of the cell to completely replicate and segregate the given chromosome. The average number of tetranucleates produced by each strain is indicated next to the key. (C) Comparative genome hybridization of total genomic DNA from wild-type (H2636, blue dots) and cdc6-mn (H2655, orange dots) cells vs a G1 DNA control (H1785) after 8 hrs in SPO. (D) Indirect immunofluorescence of Rad51 to mark DSBs and Zip1 to mark synaptonemal complex (SC) formation on spread nuclei from wild-type (H2636) and cdc6-mn (H2655) cells. Representative nuclear spreads at the 5-hr time point are shown. (E) Quantification of the number of cells showing full SC formation at the indicated times after inoculation into SPO for the strains shown in (D). The defect in SC formation is relatively mild, which is probably why it was not observed in a previous study (Brar et al., 2009). 200 cells were counted for each condition.
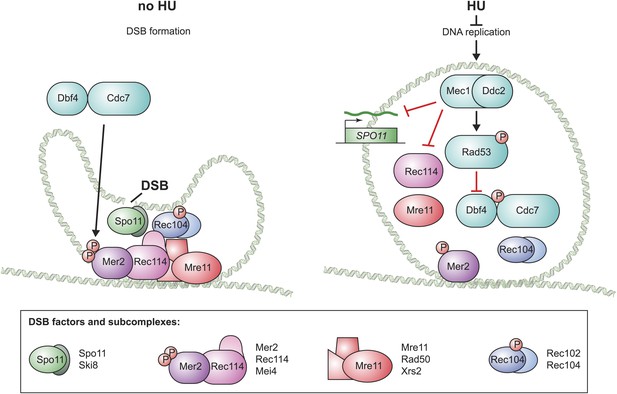
Model for the pre-meiotic replication checkpoint in budding yeast.
A schematic for the assembly of the four budding yeast DSB factor complexes (as defined below) in the absence (left) and presence (right) of replication inhibition is shown. In the absence of inhibition, all factors load onto the DNA and Mer2 and Rec104 are fully phosphorylated, allowing Spo11 to introduce DSBs. In the presence of HU, the levels of SPO11 transcripts are reduced, the DNA loading of Mre11 and Rec114 is prevented and the phosphorylation of Mer2 and Rec104 is abrogated. Illustration by Tom DiCesare (Whitehead Institute).
Additional files
-
Supplementary file 1
Genotypes of yeast strains used in this study.
- https://doi.org/10.7554/eLife.00844.016





