A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record published
- Accepted
- Received
Decision letter
-
Detlef WeigelReviewing Editor; Max Planck Institute for Developmental Biology, Germany
eLife posts the editorial decision letter and author response on a selection of the published articles (subject to the approval of the authors). An edited version of the letter sent to the authors after peer review is shown, indicating the substantive concerns or comments; minor concerns are not usually shown. Reviewers have the opportunity to discuss the decision before the letter is sent (see review process). Similarly, the author response typically shows only responses to the major concerns raised by the reviewers.
Thank you for sending your work entitled “A deletion polymorphism in the C. elegans RIG-I homolog disables viral RNA dicing and antiviral immunity” for consideration at eLife. Your article has been favorably evaluated by a Senior editor (Detlef Weigel) and 2 reviewers.
The Senior editor has assembled the following comments to help you prepare a revised submission.
Ashe and colleagues use an appealing mix of quantitative and molecular genetics to identify and characterize a natural polymorphism in the C. elegans DRH-1 gene that affects production of viral-derived primary siRNAs. The gene emerged from an analysis of natural variation in sensitivity to the Orsay virus, and by a combination of GWAS, introgression mapping, and whole-genome sequencing, the authors managed to pin down a small deletion as a major-effect locus. Using mostly if not exclusively genetics, the authors provide evidence that DRH-1 is likely to specifically recruit the Dicer protein DCR-1 to viral dsRNA substrates. This clearly is a new finding, especially in an organism with a single Dicer. While we find the work very interesting, a minimal set of additional experiments would help support the inferences of the genetic analyses by answering the following questions: 1) Does viral infection affect the expression and subcellular location of DRH-1 protein, as assayed with a GFP fusion?
2) Is there evidence that EXO RNAi is enhanced in drh-1 mutants because absence of DRH-1-bound Dicer would increase Dicer pools? That DRH-1 is not involved in endosiRNA production is consistent with a specific role for SRH-3 in bringing Dicer onto endosiRNA substrates. If the answer to the initial question is yes, this could support the scenario for the possible benefit of the drh-1 deletion in a significant fraction of the wild accessions because of enhanced environmental RNAi (which may confer unknown fitness advantages).
3) A critical experiment for supporting the proposed model would be whether the naturally found truncated form of DRH-1 indeed fails to interact with DCR-1 and RDE-4. We understand that antibodies are not available to test this. Thus, mechanistic inferences about DRH-1 action should be toned down in the revision.
https://doi.org/10.7554/eLife.00994.021Author response
1) Does viral infection affect the expression and subcellular location of DRH-1 protein, as assayed with a GFP fusion?
With regard to changes in drh-1 mRNA levels we have recently published a study analysing gene expression changes upon Orsay virus infection (Sarkies et al., Genome Research 2013). We observed no statistically significant changes in drh-1 transcript levels upon infection in wild-type (N2) animals. Thus drh-1 is unlikely to be transcriptionally regulated upon viral infection.
With regards to the subcellular localization of DRH-1 protein, we agree with the reviewers that the cell biology of viral infection in C. elegans is of great interest, including changes in the subcellular localization of DRH-1 before and after viral infection. However, we do not agree that this analysis relates directly to the manuscript that we have submitted: any change or lack of change in subcellular localization would neither strengthen nor contradict our model. Furthermore, we believe that a thorough analysis of DRH-1 behaviour upon infection would have to include analysis of the subcellular dynamics of the Orsay virus in order to be meaningful. Since we do not fully understand this yet, a combined analysis should be the focus of an independent future study. Nevertheless, we have generated a transgenic line expressing a DRH-1-GFP fusion protein. We found that these transgenic animals localize GFP diffusely in the cytoplasm of intestinal and other cells (Author response image 1 below, transgene driven by a “ubiquitous” promoter, sur-5). We observed that the cytoplasmic localization of GFP does not change in response to infection with the Orsay virus. However, in the absence of a suitable antibody that works in immunofluorescence, we are unable to assess the localization of the endogenous protein, thus we cannot rule out that changes in DRH-1 subcellular localization occur upon infection. We should therefore be cautious about the interpretation of such a negative result and we would prefer not to include these data in the manuscript. We have therefore added a paragraph to the Discussion referencing the gene expression paper, and we have referred to the results of the DRH-1GFP fusion protein analysis as data not shown.
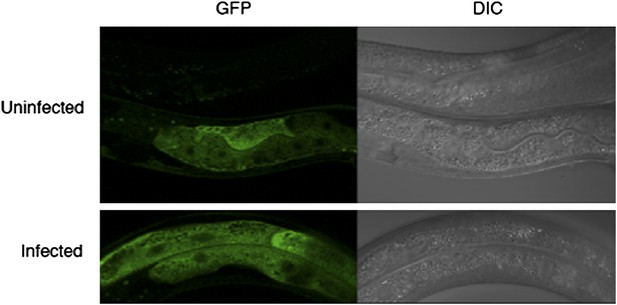
Assessing the subcellular localization of GFP in drh-1 mutant animals carrying a sur-5::drh-1::gfp transgene as an extra-chromosomal array.
Infections were carried out as described in Materials and methods. As the transgene is extrachromosomal, it is lost in some animals (top animal, top panel). We do not observe a change in GFP intensity or localization upon infection with the Orsay virus.
2) Is there evidence that EXO RNAi is enhanced in drh-1 mutants because absence of DRH-1-bound Dicer would increase Dicer pools? That DRH-1 is not involved in endosiRNA production is consistent with a specific role for DRH-3 in bringing Dicer onto endosiRNA substrates. If the answer to the initial question is yes, this could support the scenario for the possible benefit of the drh-1 deletion in a significant fraction of the wild accessions because of enhanced environmental RNAi (which may confer unknown fitness advantages).
The reviewers have raised an interesting point. We therefore performed RNAi experiments using a dilution series of unc-22 RNAi (see Materials and methods) and found that exo-RNAi is not enhanced in drh-1 mutants, while it is enhanced in eri-1 mutants. We conclude that there is no evidence that DCR-1 interaction with RDE-4 and DRH-1 limits its availability for exo-RNAi. We have included this data as Figure 4–figure supplement 4. We also added a discussion point to the Discussion.
3) A critical experiment for supporting the proposed model would be whether the naturally found truncated form of DRH-1 indeed fails to interact with DCR-1 and RDE-4. We understand that antibodies are not available to test this. Thus, mechanistic inferences about DRH-1 action should be toned down in the revision.
The truncated DRH-1 (nDf250) might fail to interact with RDE-4/DRH-1, the Orsay RNA, or both. By analogy to RIG-I we suggest that DRH-1 (niDf250) fails to recognize the Orsay RNA. However, we agree with the reviewers: we have toned down our mechanistic inferences by suggesting that DRH-1 may be necessary for either recognition of viral RNA or correct Dicer processing “assisting DCR-1 processing of the double-stranded viral RNA.”
https://doi.org/10.7554/eLife.00994.022




