ABC transporter functions as a pacemaker for sequestration of plant glucosides in leaf beetles
Figures
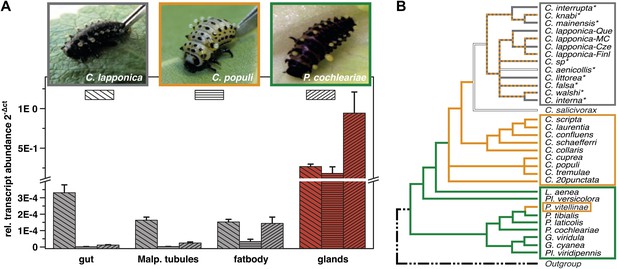
Glandular tissue-specific transcript level of cpmrp and its homologous sequences.
(A) Relative transcript abundance of cpmrp (C. populi) and its homologous sequences from C. lapponica and P. cochleariae in different larval tissues (n = 3–4, mean ± SD) assigned to (B) their phylogenetic group and chemical defense strategies based on maximum parsimony reconstruction (according to Termonia et al., 2001). Green, autogenous group of monoterpene iridoid producers; orange, obligate-sequestering group; gray, interrupta group with mixed metabolism that evolved the biosynthesis of butyrate-esters.
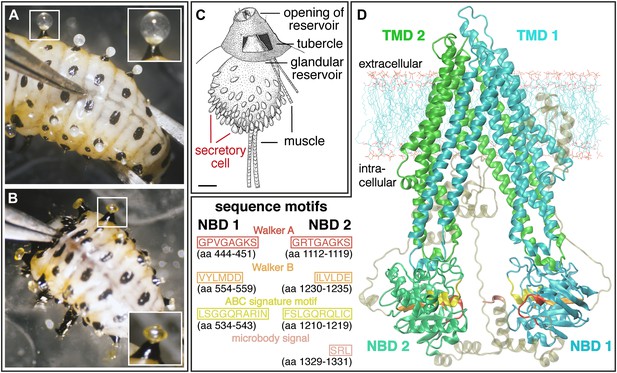
Silencing effect and 3D-structure model of CpMRP.
(A and B) Production of defensive secretions is disrupted in CpMRP knockdown L3 larvae (B) compared to the phenotype of the control larvae (A). (C) Drawing of dissected glandular tissue of C. populi according to Hinton, 1951 with relaxed reservoir in contrast to the everted reservoir in insets of (A) and (B). (D) 3D-model of CpMRP, embedded in a lipid bilayer, illustrating its probable correct global topology based on I-TASSER (TM-score of 0.52 ± 0.15; C-score: −1,57) and the localization of characteristic sequence motifs.
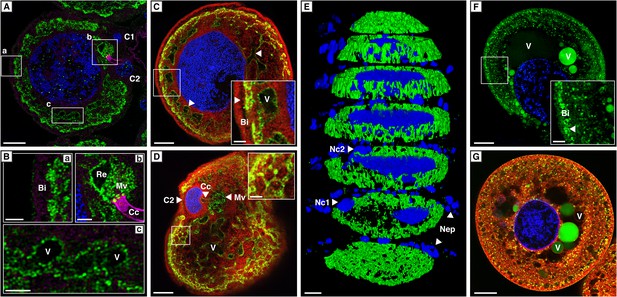
Localization of CpMRP in secretory cells of C. populi.
(A–G) High-magnification optical sections through secretory cells. (A–E) Immunohistochemical staining of CpMRP (green) in fixed secretory cells. CpMRP staining was confined to intracellular Bodipy-stained membrane structures and displayed a distinct reticular pattern. (B) Extracted cutouts of (A and C) optical section through the nucleus, (D) optical section above the nucleus of the secretory cell, (E) 3D stack displaying CpMRPs primarily spherical distribution. (F and G) CDCFDA staining for vacuolar esterase activity (green) in live cells. A multitude of vacuoles are present that vary in their enzyme content. Bi = basal infoldings, C1 and C2 = canal cells, Cc = cuticular canal, Mv = microvilli, Nc1, Nc2 = nucleus of canal cells, Nep = nuclei of epithelium cells, Re = extracellular room, V = vacuole, Blue, nuclear staining; Red, Bodipy-stained intracellular membrane; Magenta, false color-coded autofluorescence. Scale bars, 20 µm or 5 µm (insets).

Localization of CpMRP in whole larvae cryosections of C. populi.
Overlay of brightfield and immunofluorescence images of lateral cryosection of an entire L2 stage larvae of C. populi. Green, CpMRP; Blue, nuclear staining. The black inset depicts an orientation overview of the cut larvae. The red inset shows an exemplary secretory cell with higher magnification. Scale bars, 500 µm or 50 µm (red inset).
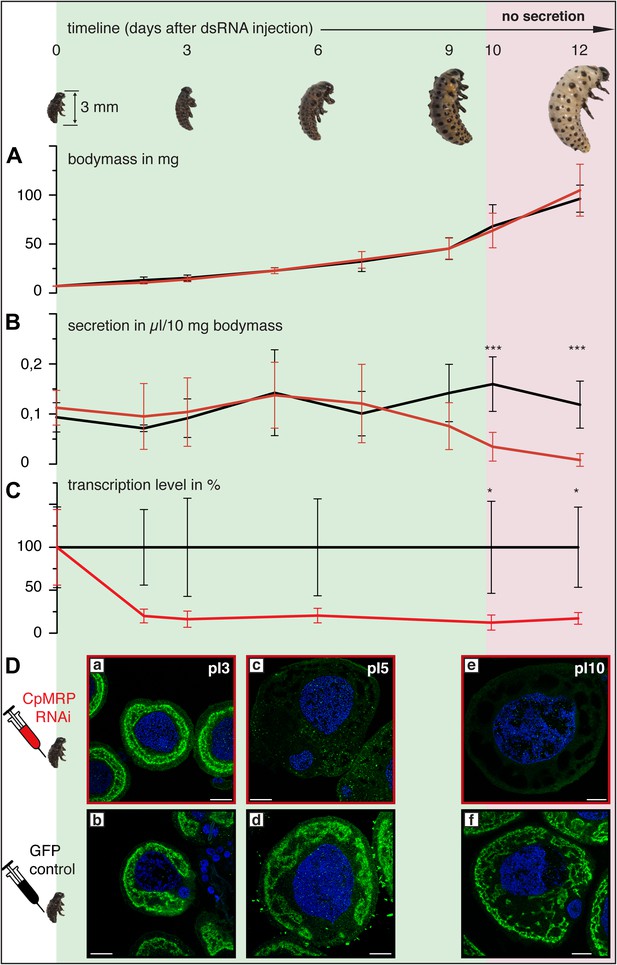
Timeline of different CpMRP knockdown effects.
(A–D) C. populi larvae development following CpMRP knockdown by dsRNA injection into the larval hemocoel (d 0). CpMRP knockdown effects; red/(D a, c, e) were compared to the gfp-injected control larvae; black/(D b, d, f) at different developmental stages. (A) Larval fitness (body mass) was not influenced by CpMRP knockdown (n > 10, mean ± SD). (B) CpMRP knockdown larvae lack defensive secretions 10 days after dsRNA injection (n > 10, mean ± SD). (C) Transcriptional level of cpmrp inside the glands (each time point contains n = 3 (biological replicates), mean ± SD). (D) CpMRP protein level decreased after dsRNA injection—Green, CpMRP; Blue, nuclear stain. Scale bars, 20 µm; pIx = x days post dsRNA-injection. Asterisks represent significant differences in cpmrp-silenced larvae compared to gfp-injected control larvae (*p≤0.05, ***p≤0.001).
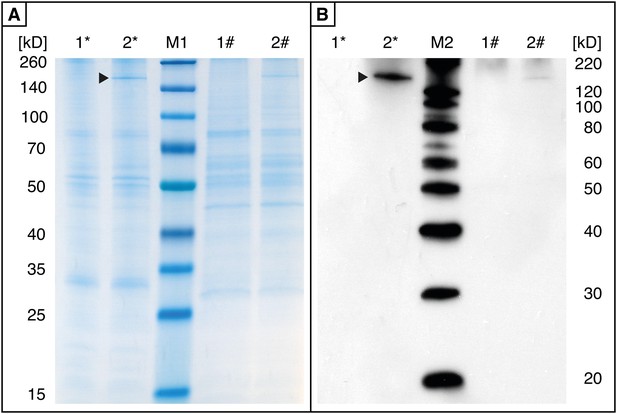
Effects of cpmrp silencing on the protein level of the glandular tissue of C. populi.
(A and B) Total protein analysis of glandular tissue from cpmrp-knock-down (1) and gfp-control larvae (2) of C. populi (10 days post dsRNA-injection). 5 µg total protein contents of roughly separated membrane protein fraction (*) or cytosolic proteins (#) were separated by SDS-PAGE; (A) Coomassie staining; (B) Western blot with anti-CpMRP. Arrows indicate CpMRP in the membrane protein fraction.
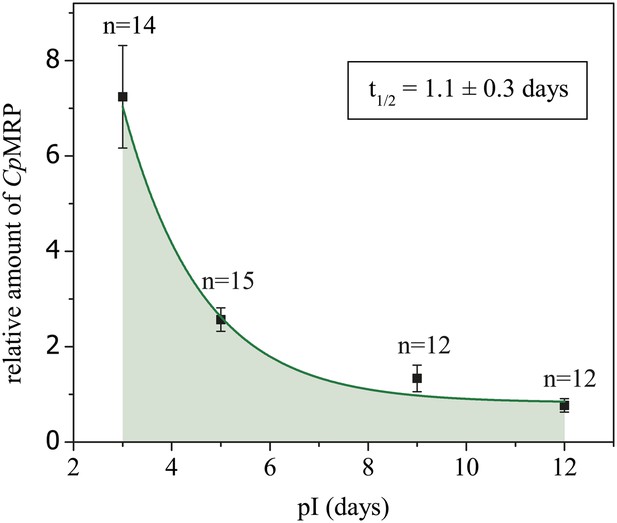
Degradation kinetics of CpMRP in secretory cells of C. populi.
Immunohistochemical staining was employed to follow the degradation kinetics of CpMRP with Alexa 488 after cpmrp dsRNA injection (d 0). Integrated Alexa 488 fluorescence intensity from secretory cells was normalized to autofluorescence.

DNA alignment of cpmrp–related ABC transporter sequences in C. populi.
Alignment (ClustalW) of cpmrp and 7 closely related ABC transporter sequences in C. populi (black = exact match with cpmrp). Amino acid sequence identity of CpMRP to transporter OT-1 (76.1%), OT-2 (69.4%), OT-3 (61.6%), OT-4 (58.3%), OT-5 (63.9%), OT-6 (64.9%) and OT-7 (64.1%); OT = potential off-target.
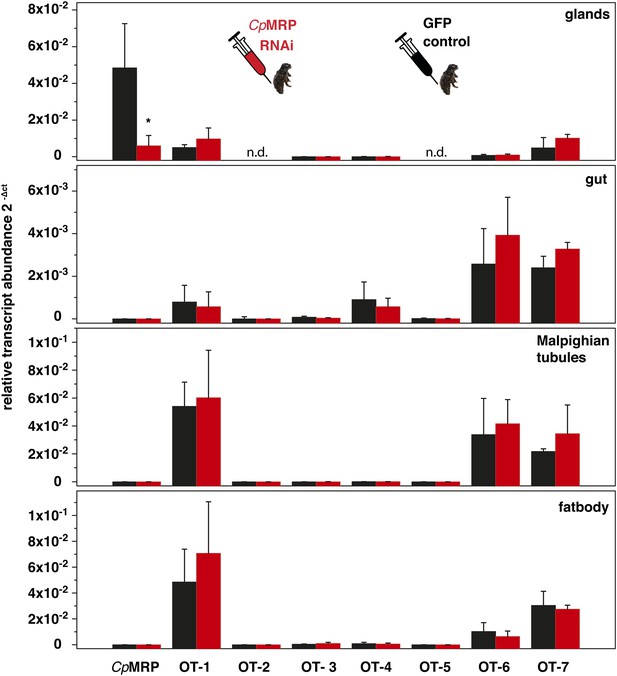
Evaluation of possible off-target effects of CpMRP dsRNA in larval tissue of C. populi.
Relative transcript abundance (2ΔCt) of cpmrp and closely related possible off-target ABC transporter sequences (OT 1–7) in different larval tissues 10 days after dsRNA injection (n = 4, mean ± SD). Transcript abundance in cpmrp-dsRNA (red) was compared to gfp-injected control larvae (black). CpActin was used for normalization of transcript quantities. Asterisks indicate significant differences between gfp-injected control larvae and cpmrp-silenced larvae (*p≤0.05), n.d.=not detectable.
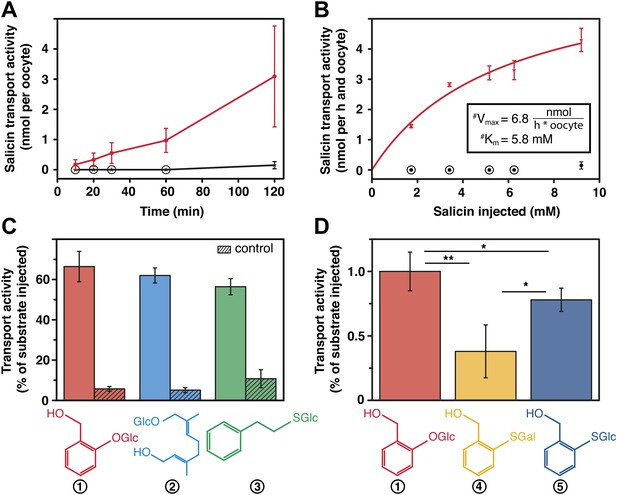
Salicin transport activity of CpMRP in Xenopus laevis oocytes.
(A–D) Transport activity was determined by quantifying the substrate efflux in the oocyte incubation medium of cpmrp-RNA compared to water-injected control oocytes via HPLC-MS. (A) Time course of CpMRP-dependent salicin efflux after the injection of 5 nmol salicin (incubation time: 1 hr, n = 5, mean ± SD). Red, CpMRP-expressing oocytes; Black, water-injected control. (B) Concentration dependence of CpMRP-mediated salicin transport (red); water-injected control in black (n = 5, mean ± SD, #: apparent, encircled data point: not detectable). (C) Comparative transport assays of CpMRP activity with a substrate mixture (salicin (1), 8-hydroxygeraniol-O-glucoside (2) and phenylethyl-S-glucoside (3)). Open bar, transport activity of CpMRP-expressing oocytes. Crosshatched bar, transport activity water-injected control oocytes. (D) Comparative transport assays of CpMRP salicin transport activity to thiosalicin (5) and its galactoside analogue (4) and thiosalicin (incubation time: 1 hr, n = 10, mean ± SD). Asterisks represent significant differences among indicated substrates (*p≤0.05, **p≤0.01). Encircled data points represent undetectable concentrations.
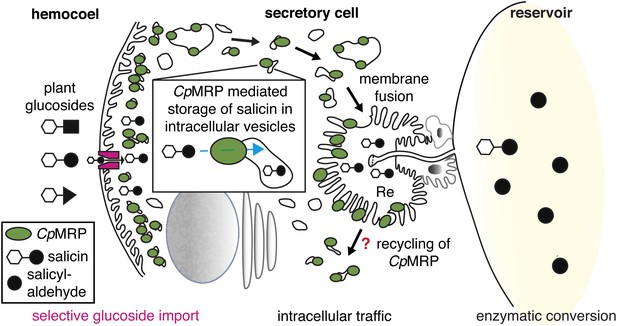
CpMRPs pacemaker function and sequestration model.
Schematic view of our sequestration model through a secretory cell (see Figure 2C: overview of the defensive system, secretory cells are indicated in red). Different plant glucosides (black triangles, circles and squares joined to a glucose molecule indicated by a white hexagon) circulate in the hemocoel. CpMRP dictates (pacemaker function) the transport rate of a still unknown selective, maybe gradient-driven transporter (magenta) for salicin in the plasma membrane by a constant accumulation of salicin in intracellular vesicles. These vesicles are tracked via exocytosis to the reservoir where the enzymatic conversion of salicin to salicylaldehyde takes place.
Videos
3D representation of CpMRP localization within a secretory cell.
Exterior view of a secretory cell of C. populi based on immunohistochemical staining (green, CpMRP; blue, nuclei stain). The z-stack was acquired with a resolution of x = 0.146 µm; y = 0.146 µm and z = 0.500 µm. The smallest dimension of the depicted secretory cell is about 100 µm.
3D representation of CpMRP localization within a secretory cell.
Interior view of Video 1. The camera is centrally positioned within the nucleus and rotates within the plane of the first frame of Video 1, initially pointing to the area between the nuclei of the two canal cells.
Tables
Oligonucleotide primers
| Gene name | Primer name | cpmrp RACE |
|---|---|---|
| cpmrp Gene-Bank: KC112554 | 3´RACE | GCACGGTCCTGACTATAGCGCACAGGC |
| 5´RACE | CCTGCCCCCGTTCTTCCCACAATACC | |
| 2nd 5´RACE | GGTGGAGGCCTGCATGGTCAGCTTGC | |
| 5´nested | CGGCGTCTCGAATGGACCTTCCGTGTCG | |
| 3´nested | GGAGAGATGGTGGGAGTATGACCACCCC |
| Gene name | Primer name | Primer for ds RNA generation |
|---|---|---|
| cpmrp Gene-Bank: KC112554 | fwd | GATTAATACGACTCACTATAGGCGACTAAGTGTGAACTAGTCGGTGC |
| rev | GATTAATACGACTCACTATAGGGAGACTTGTCTCCACAGCAGATAG | |
| gfp UniProtKB: P42212.1 | fwd | TAATACGACTCACTATAGGGAGATGGCTAGTAAGGGA |
| rev | TAATACGACTCACTATAGGGAGATTATTTGTAGAGTTC |
| Gene name | Primer name | qPCR Primer |
|---|---|---|
| cpRP-L45 GeneBank: JX 122918 | fwd | CACTGGAATCCAAAGTGGAAACTG |
| rev | CTGCCTTTCAACCCATGGTC | |
| cpActin GeneBank: JX122919 | fwd | ACGTGGACATCAGGAAGGAC |
| rev | ACATCTGCTGGAAGGTGGAC | |
| pcRP-L8 Gene-Bank: JX122920 | fwd | CATGCCTGAAGGTACTATAGTGTG |
| rev | GCAATGACAGTGGCATAGTTACC | |
| cpmrp Gene-Bank: KC112554 | fwd | CCTGGATCCATTCGATGAGT |
| rev | AGTATCGCCCTCGCTAGACA | |
| pcmrp Gene-Bank: KF278996 | fwd | CTCTAGACATCATGGTCACAGA |
| rev | GGCATATCAACTGTCGTTGTC | |
| clapmrp Gene-Bank: KF278997 | fwd | CCATCTGGCAAATTTGAAGATTTC |
| rev | AGTATTGCCCTCGCTAGACA | |
| off-target_OT-1 | fwd | GGAATTCGAGGACCAGTTGC |
| rev | GGCATATCAACTGTCGTTGTC | |
| off-target_OT-2 | fwd | CCATCTGGCAAATTTGAAGATTTC |
| rev | CAGTAACTACGAAGTCTAGAGAG | |
| off-target_OT-3 | fwd | GAATATCAGATGCCGACCTG |
| rev | GGATGGCTCTGGCAAGG | |
| off-target_OT-4 | fwd | GACGACGTGCTTTATAGAGC |
| rev | GTCCCACGCTATAGTTGGAC | |
| off-target_OT-5 | fwd | CAGTATTCGTTATAACTTGGACC |
| rev | GTCCTGCGCTGAAATTCGATC | |
| off-target_OT-6 | fwd | GACGAATATAAGGATGAGACATTG |
| rev | GCGTACTATGGCTCGAGC | |
| off-target_OT-7 | fwd | AGCCGATGATGCAACCATC |
| rev | CCAACTGTCTTTGTCCAGC |
-
All primers are listed by name, sequences and application. Forward primers are indicated as ‘fwd’ and reverse primers as ‘rev’.





