MEGF8 is a modifier of BMP signaling in trigeminal sensory neurons
Figures
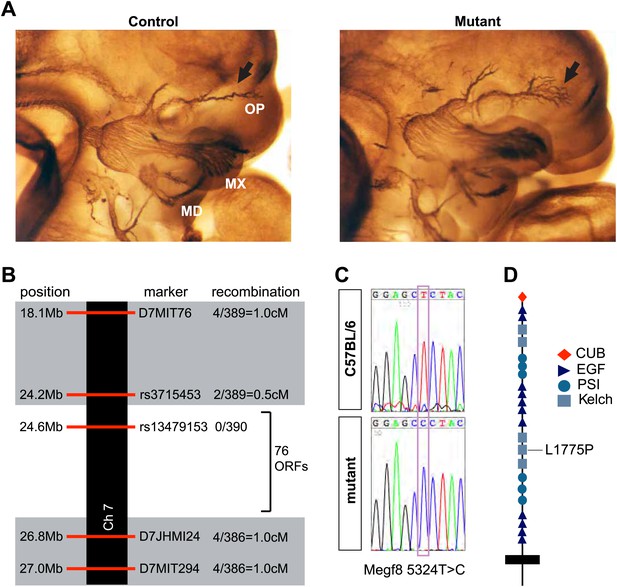
Disruption of Megf8 causes defasciculation of the TG ophthalmic nerve.
(A) Whole-mount neurofilament staining of E11.5 control and Line 687 mutant littermates, showing the trigeminal ganglia (TG) and its three main projections: the ophthalmic (OP), maxillary (MX), and mandibular (MD) branches. (B) Schematic diagram of the region of Chromosome seven found to contain the Line 687 mutation, the markers used to diagnose linkage, and the frequency of recombination events observed at these markers. ORF, open reading frame. (C) Sequence data highlighting the mutation (Megf8 5324T>C) observed in Line 687 mutant DNA compared with C57BL/6 wild-type DNA. (D) Schematic diagram of Megf8. The Line 687 point mutation induces a single amino acid substitution L1775P in a Kelch domain of Megf8.
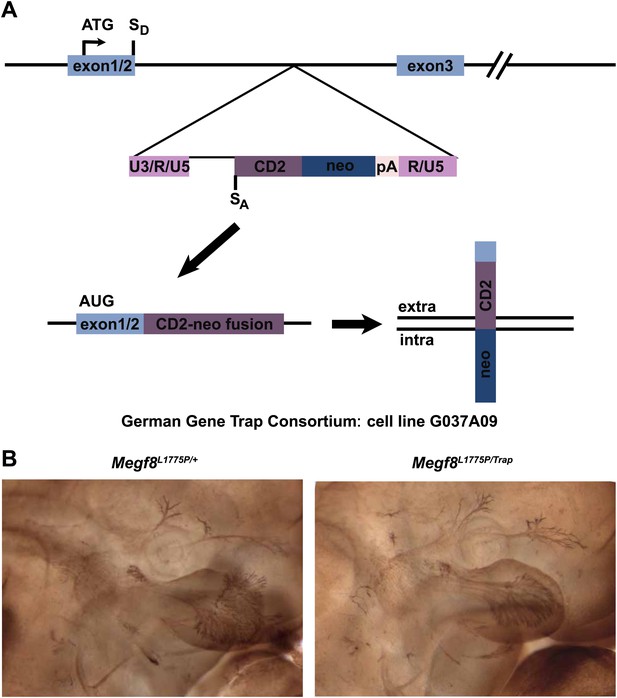
Complementation analysis of Megf8Trap and Megf8L1775P alleles.
(A) Schematic diagram of gene trap allele (Megf8Trap). German Gene Trap Consortium cells G037A09 have an intron 2–3 insertion of a secretory trap vector such that the start ATG and signal peptide of Megf8 is captured and fused to a CD2-neomycin fusion protein instead of the remaining endogenous locus. SD = splice donor, SA = splice acceptor. (B) Whole-mount neurofilament staining of E11.5 embryos from an intercross of Megf8Trap/+ and Megf8L1775P/+ heterozygotes. Resulting Megf8L1775P/Trap embryos have the trigeminal defasciculation phenotype.
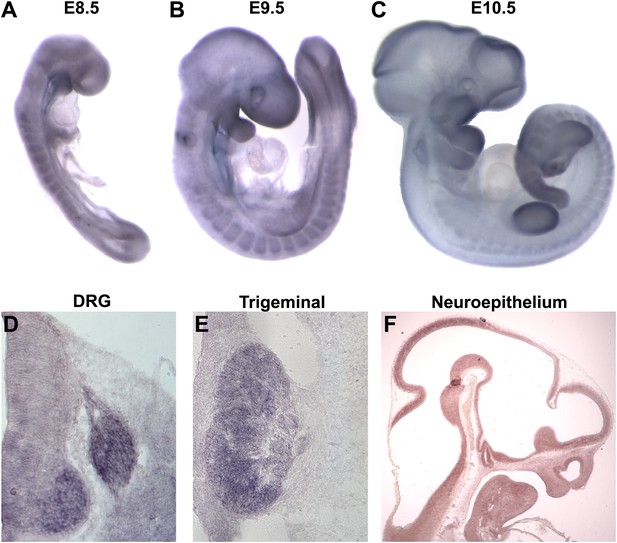
Megf8 is expressed widely during development.
(A–C) Whole-mount in situ hybridization (ISH) for Megf8 at E8.5, E9.5, and E10.5. (D) ISH for Megf8 on E11.5 transverse cryosection shows expression in the DRG. (E) ISH for Megf8 on E11.5 transverse cryosection shows expression in the TG. (F) ISH for Megf8 on E10.5 paraffin sagittal section shows expression in the developing neuroepithelium.
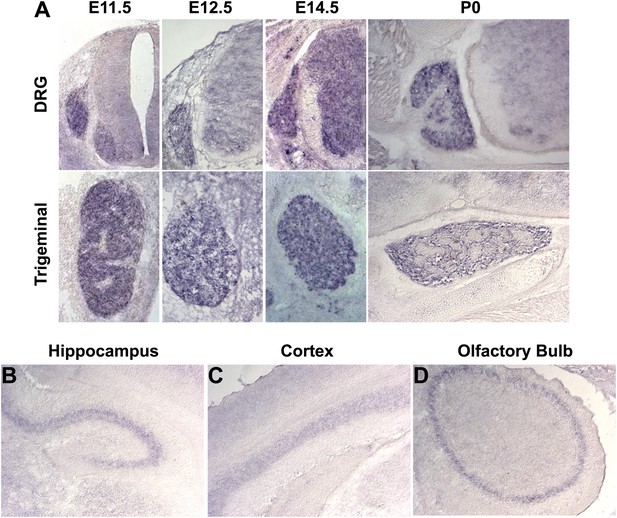
Megf8 is expressed throughout the developing nervous system.
(A) In situ hybridization on coronal cryosections from E11.5-P0 shows strong Megf8 expression at all time points in the DRG and TG. (B–D) In situ hybridization on coronal cryosections at P0 shows strong Megf8 expression in the hippocampus (B), layer 4/5 of the cortex (C), and in the olfactory bulb (D).
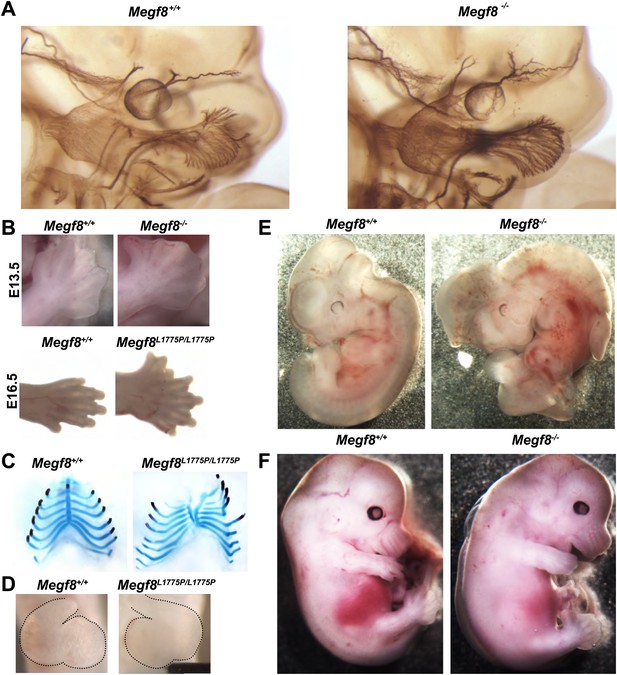
Megf8 is required for development of the trigeminal ganglia, limb, skeleton, heart, and left-right asymmetry.
(A) Whole-mount neurofilament staining of E11.5 Megf8+/+ and Megf8−/− littermates. The Megf8−/− null mutant phenocopies the point mutant Megf8L1775P/L1775P. (B) Whole-mount images of Megf8+/+ and Megf8−/− hindlimbs at E13.5 (top) and Megf8+/+ and Megf8L1775P/L1775P forelimbs at E16.5 (bottom). (C) Alcian blue and alizarin red staining of E16.5 embryonic ribs/sternum. Megf8L1775P/L1775P mutants have a split sternum and delayed ossification of the rib cage. (D) Whole-mount images of the heart of freshly fixed E10.5 embryos with dissected pericardial cavity. Megf8L1775P/L1775P have complete left-right inversion of heart looping. Heart is outlined with dotted lines. (E) Whole-mount images of E11.5 Megf8+/+ and Megf8−/− littermates, showing reversal of embryonic turning and exencephaly in the Megf8−/−. (F) Whole-mount images of Megf8+/+ and Megf8−/− littermates at E13.5, showing severe edema in the Megf8−/−.
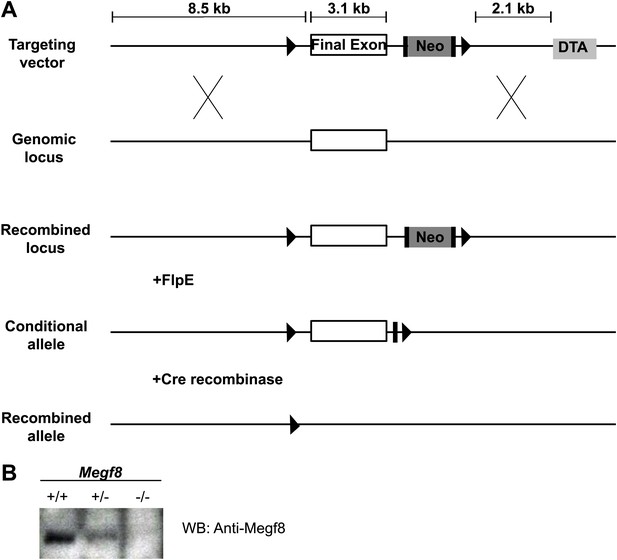
Generation of a conditional knock-out mouse line (Megf8Flox).
(A) Schematic diagram of Megf8 gene targeting strategy. This strategy targeted the final exon of Megf8, which is 3.1 kb and comprises the transmembrane domain, intracellular C-terminus, and 3’′ UTR. A targeting vector was designed with an 8.5 kb long arm, a single loxP site located 500 bp upstream of the targeted exon, a frt-Neo-frt-loxP cassette placed 500 bp downstream of the exon, a 2.1 kb short arm, and a DTA selection cassette. Recombination with the endogenous locus resulted in integration of the 5′ loxP and 3′ frt-Neo-frt-loxP sites. Following germline transmission, male carriers were then mated with FlpE females to excise the neomycin cassette and generate the conditional allele. (B) Western blot of E12.5 TG and DRG lysates from Megf8+/+, Megf8+/−, and Megf8−/− littermates. The blot was probed with rabbit anti-Megf8 and shows a loss of Megf8 protein in Megf8−/− neurons.
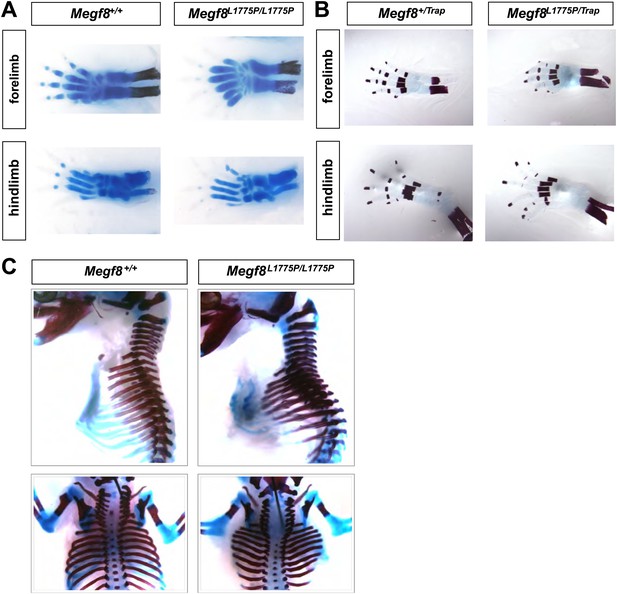
Megf8L1775P/L1775P mutants show defects in limb and skeletal development.
(A) Alcian blue staining on E16.5 limbs shows that Megf8L1775P/L1775P mutants have digit duplication as well as duplication of bones in the hand. (B) Alizarin red staining on E16.5 limbs shows that Megf8L1775P/Trap mutants have duplication of the bones of the autopodium. (C) Alcian blue and alizarin red staining of whole E16.5 embryo skeletons. Megf8L1775P/L1775P mutants have a wider and shorter ribcage, delayed ossification of the ribcage, and a split sternum.
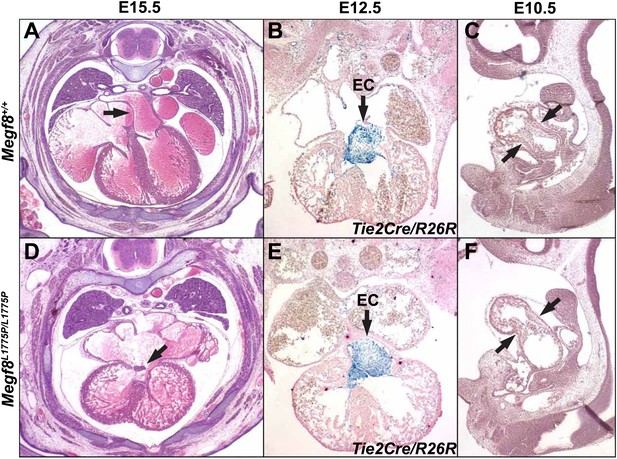
Heart development in Megf8L1775P/L1775P mutant embryos.
Sections from control (A–C) and Megf8L1775P/L1775P mutant (D–F) embryos. (A and E) Hematoxylin and eosin (H and E) staining of E15.5 control and mutant sections. The arrowhead in the control E15.5 embryo section points to the atrial septum; the arrowhead in the mutant embryo section points to the vestige of the endocardial cushion, above which the atrial septum is missing and below which the atrioventricular valves are poorly formed or atretic. (B and E) β-galactosidase and nuclear fast red staining of E12.5 embryos, which also carried the Tie2Cre/R26R alleles for labeling endothelium and endocardium and derived mesenchymal cells (Kisanuki et al., 2001; Soriano, 1999). Note the normal endocardial cushion (EC) mesenchyme (arrows). (C and F) H and E staining of E10.5 control and mutant sections. Note the apparently normal atrioventricular cushion mesenchyme at E10.5 (arrows).
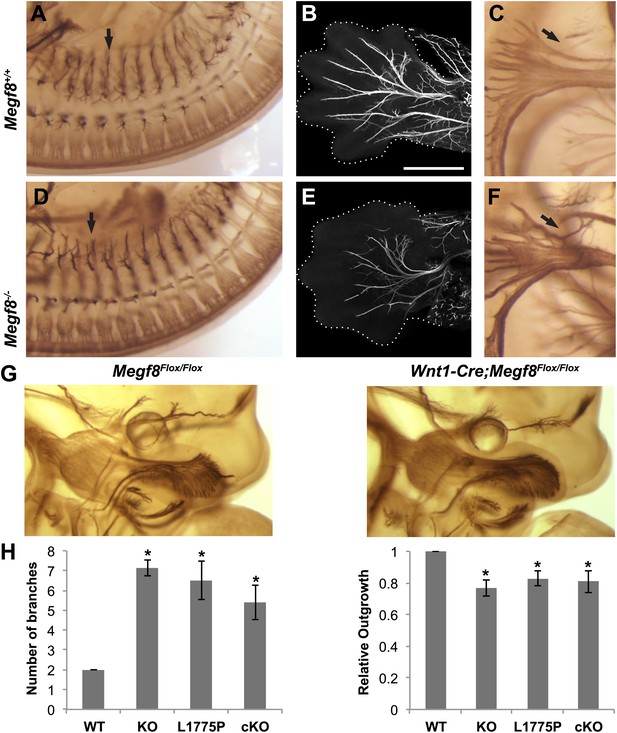
Megf8 is required for development of the PNS.
(A and D) Whole-mount neurofilament staining of E11.5 Megf8+/+ and Megf8−/− littermates showing the DRG spinal nerves, which are undergrown in the Megf8−/− (arrow). (B and E) Whole-mount peripherin staining of forelimbs from E13.5 Megf8+/+ and Megf8−/− littermates. The radial and ulnar nerves are undergrown in the Megf8−/−embryo. Limbs are outlined with dotted lines. Scale bar (B and E) represents 500 μm. (C and F) Whole-mount neurofilament staining of E11.5 Megf8+/+ and Megf8−/− littermates. The vagus/glossopharyngeal nerves are defasciculated in the Megf8−/− (arrow). (G) Whole-mount neurofilament staining of E11.5 Megf8Flox/Flox and Wnt1-Cre; Megf8Flox/Flox littermates. (H) Quantification of ophthalmic branch phenotype for Megf8−/− (KO), Megf8L1775P/L1775P (L1775P), and Wnt1-Cre; Megf8Flox/Flox (cKO) compared to Megf8+/+ (WT). Left: the number of branches at the nasociliary branch point was significantly greater for KO, L1775P, and cKO embryos. Right: the ophthalmic branch was undergrown in KO, L1775P, and cKO embryos. Four to seven embryos were analyzed per genotype. Error bars represent mean ± s.e.m. *p<0.05, one-way ANOVA. The relative outgrowth was also measured for the maxillary and mandibular branches of the TG for Megf8−/− embryos (not shown). The maxillary branch was slightly undergrown compared to Megf8+/+ (relative outgrowth 0.9, p<0.05) while the mandibular branch was unaffected (relative outgrowth 0.99, p=0.7). To assess defasciculation in the maxillary branch, the relative maxillary area was calculated and no difference was observed between Megf8+/+ and Megf8−/− (relative area 0.94, p=0.2).
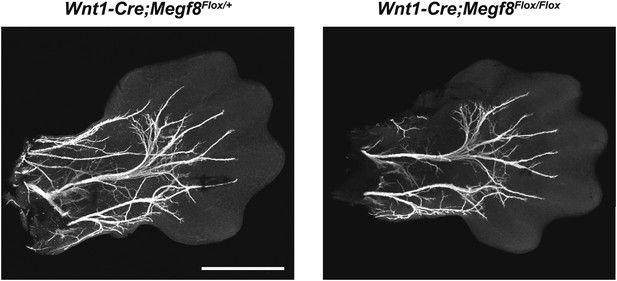
Conditional deletion of Megf8 from DRG neurons does not disrupt formation of the radial/ulnar nerves.
Whole-mount peripherin staining of E13.5 forelimbs from Wnt1-Cre;Megf8Flox/+ and Wnt1-Cre;Megf8Flox/Flox littermates. Limbs are outlined with dotted lines. Scale bar represents 500 µm.
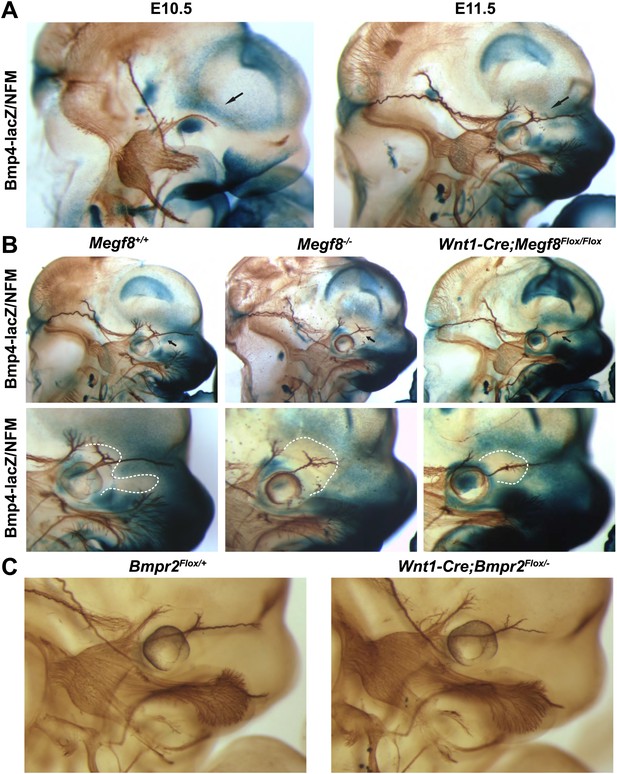
BMP signaling is required for proper extension of the TG ophthalmic nerve.
(A) Whole-mount β-galactosidase and neurofilament (NFM) co-staining on Bmp4lacZ/+ embryos at E10.5 and E11.5, showing the relationship between Bmp4 expression and the developing TG nerve. (B) Whole-mount β-galactosidase and neurofilament co-staining on E11.5 embryos: Bmp4lacZ/+;Megf8+/+ (left) and Bmp4lacZ/+;Megf8−/− (center) littermates, as well as Bmp4lacZ/+;Wnt1-Cre/Megf8Flox/Flox (right). Bmp4lacZ expression is lost at the location where defasciculation of the TG ophthalmic nerve occurs in Megf8−/− and Wnt1-Cre;Megf8Flox/Flox embryos. The top row shows the whole head with all three TG branches. The bottom row is an enlargement of the top row showing the ophthalmic branch. The area of perturbed Bmp4 expression is outlined. The disruption of Bmp4lacZ expression was fully penetrant and observed in all Megf8−/− (n = 4) and Wnt1-Cre;Megf8Flox/Flox (n = 3) embryos assessed. (C) Whole-mount neurofilament staining of E11.5 Bmpr2Flox/+ and Wnt1-Cre;Bmpr2Flox/− littermates.
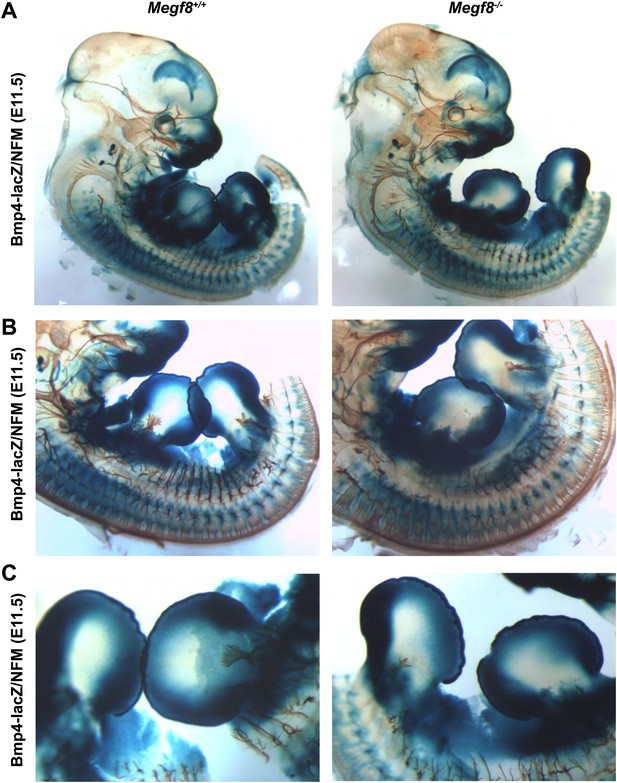
Bmp4 expression in Megf8−/− embryo compared to wild-type littermate.
Whole-mount β-galactosidase and neurofilament co-staining on E11.5 embryos: Bmp4lacZ/+;Megf8+/+ (left) and Bmp4lacZ/+;Megf8−/− (right) littermates. Bmp4lacZ expression is lost at the location where defasciculation of the TG ophthalmic nerve occurs in Megf8−/− embryos but is intact throughout the rest of the embryo. (A) Presentation of the whole embryo. (B) DRG and spinal nerves. (C) Forelimbs and hindlimbs.
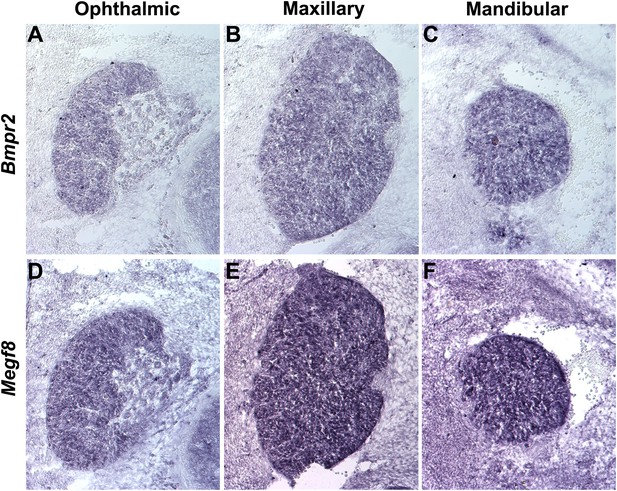
Bmpr2 and Megf8 are expressed throughout the developing TG.
In situ hybridization on serial transverse cryosections at E12.5 shows strong expression of Bmpr2 (A–C) and Megf8 (D–F) throughout the developing TG. Representative images are shown for ophthalmic (A and D), maxillary (B and E), and mandibular (C and F) lobes of the TG.
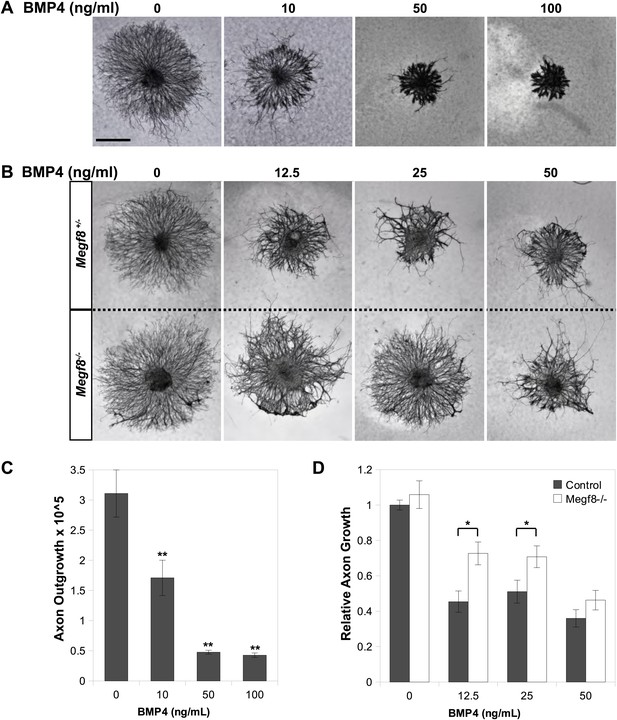
Megf8 mediates the inhibition of TG axon growth by BMP4.
(A) E12.5 wild-type TG explants cultured in 0–100 ng/ml BMP4. (B) E12.5 TG explants from Megf8+/− and Megf8−/− littermates cultured side by side in 0–50 ng/ml BMP4. (C) Quantification of (A): axonal outgrowth of wild-type TG explants in the presence of 0–100 ng/ml BMP4. Increasing doses of BMP4 caused robust inhibition of axon outgrowth (**p<0.05, Student’s t test). Three to four explants were quantified for each concentration of BMP4. Error bars represent mean ± SEM. (D) Quantification of (B): relative axon outgrowth of TG explants from control (Megf8+/+ or Megf8+/-) and Megf8-/- littermates in the presence of 0–50 ng/ml BMP4. BMP4 inhibition of axon outgrowth is partially lost in Megf8−/− explants (*p<0.05, Student’s t test). The experiment was repeated three times, using three to four explants per BMP4 concentration in each experiment; each bar represents results from at least 10 explants. Error bars represent mean ± SEM. Scale bar represents 20 μm.
Tables
Summary of Megf8−/− phenotypes and implicated BMPs
| Phenotype | Megf8-/- | BMP implicated | |
|---|---|---|---|
| Trigeminal nerve (V1) defasciculation | 100% (10/10) | n.d. | |
| Trigeminal patterning | n.d. | BMP4 | (Hodge et al., 2007) |
| Polydactyly | 100% (9/9) | BMP4, BMP7 | (Dudley et al., 1995; Dunn et al., 1997) |
| Reversed heart looping | 33% (7/21) | BMP4 | (Fujiwara et al., 2002) |
| Reversed embryonic turning (E11.5) | 33% (5/15) | BMP4 | (Fujiwara et al., 2002) |
| Edema (E13.5+) | 100% (6/6) | ||
| DRG spinal nerves undergrown (E11.5) | 100% (10/10) | BMP4 | (Guha et al., 2004) |
| Radial/ulnar nerves undergrown (E13.5) | 100% (2/2) | BMP4 | (Guha et al., 2004) |
| Vagus defasciculation | 90% (9/10) | n.d. | |
| Exencephaly | 36% (16/45) | ||
| Disrupted BMP4 expression around V1 | 100% (4/4) | ||
-
Phenotypes, stages of observation and penetrances of Megf8−/− mutants are listed. V1 refers to the ophthalmic branch of the trigeminal nerve. Bmp loss-of-function lines known to display similar phenotypes are noted.





