Chromatin signature of widespread monoallelic expression
Figures
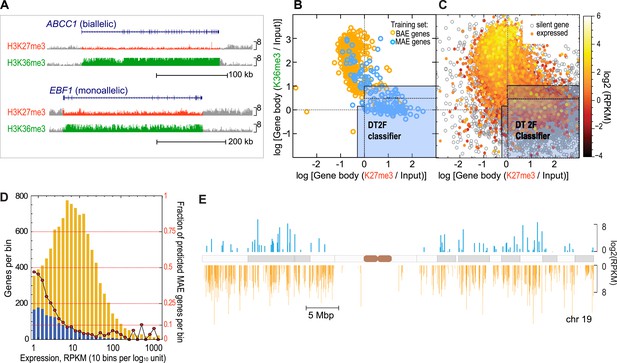
Genes with monoallelic expression have specific chromatin signature within the gene body.
(A) Assessment of histone modifications. The mapped ChIP-Seq signals for the listed modifications were derived from the total signal over the gene-body or promoter region: shown is the gene body signal for the two most informative chromatin marks H3K36me3 (green) and H3K27me3 (red). EBF1 gene was shown to be MAE, ABCC1 was shown to be biallelic in lymphoblastoid cells (Gimelbrant et al., 2007). ChIP-Seq data in GM12878 lymphoblasts were generated by the ENCODE project. Graphics adapted from UCSC genome browser (http://genome.ucsc.edu/; Meyer et al., 2013). Height of the signal tracks was set 0–8. (B) High confidence MAE (blue) and biallelic (gold) autosomal genes in the training set are separated by the gene body signal for H3K27me3 and H3K36me3 in GM12878 cells. Light blue area illustrates partitioning of this space by the most optimal classifier (DT2F). Solid line demarcates external border of ‘Neutral’ setting; dotted line shows more restrictive ‘Precision’ setting and is a graphical representation of the boundary identified by an alternating decision tree (DTree), which was the best-performing machine learning method applied to the features after feature selection. Of 270 high confidence MAE genes, 268 had data for both H3K27me3 and H3K36me3. Of these, 204 (76%) are within predicted MAE region. (C) Distribution of all autosomal RefSeq genes in GM12878 cells according to gene body signal for H3K27me3 and H3K36me3. Genes are color-mapped according to their expression level in GM12878 cells, from lowly expressed in red to highly expressed in yellow. Silent transcripts (RPKM <= 0.1) are shown in gray. Solid and dotted lines as in 1B. (D) Fraction of predicted MAE genes as a function of gene expression level. Left vertical axis: absolute number of predicted MAE (blue) and non-MAE genes (gold) per expression level bin. Right axis: fraction of predicted MAE genes (red circles) per same bin. Expression bins are 0.1 log10 units of RPKM in GM12878 cells. (E) Genome distribution of predicted MAE and biallelic genes and their expression level. Shown is chromosome 19; other autosomes are similar. Blue—genes predicted as MAE; gold—genes predicted as biallelic. Position along the chromosome corresponds to transcription start site of the gene; marker length reflects gene expression level in GM12878 cells. Only genes with RPKM > 1 are shown.
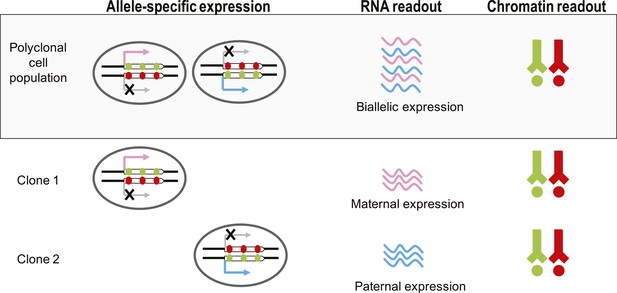
Chromatin signature of monoallelic expression allows its detection in monoclonal and polyclonal samples.
Detection of MAE by expression bias is not possible in polyclonal cell populations as both paternal and maternal transcripts are present, making expression appear biallelic. H3K36me3 is indicated by green circles and H3K27me3 is indicated by red circles.
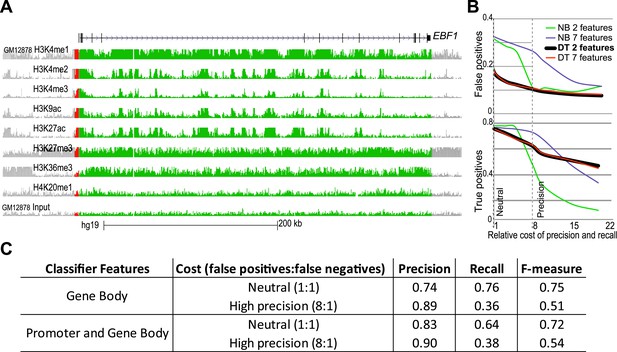
Building and performance of chromatin feature classifiers.
(A) The mapped ChIP-Seq signals for the listed modifications were derived from the total signal over the gene-body (green) or 2.5 kb promoter region (red). EBF1 gene was shown to be MAE in lymphoblastoid cells (Gimelbrant et al., 2007). ChIP-Seq data in GM12878 lymphoblasts were generated by the ENCODE project. Graphics adapted from UCSC genome browser (http://genome.ucsc.edu/; Meyer et al., 2013). Height of the signal tracks was set 0–8. (B) Comparison of precision and recall of different classifier types when using distinct sets of chromatin features. False positive (FP) and false negative (FN) calls for training set of MAE and BAE genes are shown as function of the increasing cost of false positive errors. Classifiers shown: DT–Decision Tree; NB–Naïve Bayes. Feature sets: ‘7 features’–gene body signal for H3K27me3 and H3K36me3; and promoter signal for H3K27me3, H3K36me3, H3K4me2, H4K20me3, and H3K27ac; ‘2 features (also called DT2F)’—only gene body signals for H3K27me3 and H3K36me3. Neutral and Precision settings were chosen, respectively, for best recall, and for the optimal combination of recall and precision. (C) Comparison of the 2-feature (GeneBody) and 7-feature (GenePromoterAndBody) classifiers. Similarity of precision and recall values suggests that the two chromatin marks, H3K27me3 and K3K36me3, account for most of the difference between MAE and BAE genes.
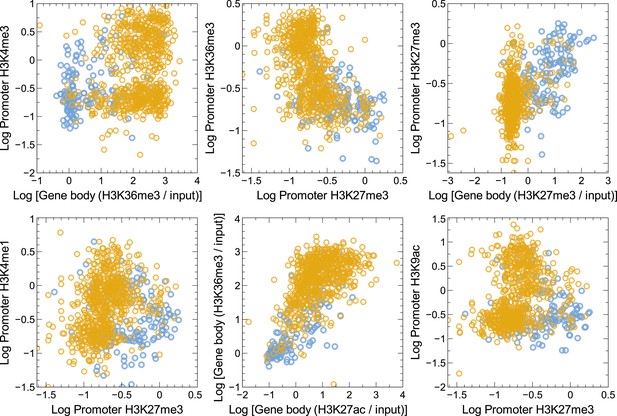
Distribution of various promoter and/or normalized gene body signal combinations in GM12878 cells in our training set.
High confidence MAE (blue) and biallelic (gold) autosomal genes in the training set do form clusters in some of these cases but fail to achieve as clear a separation of MAE and biallelic genes as does normalized H3K27me3 and H3K36me3 gene body signal combination (Figure 1B). Data are shown for a few representative cases.
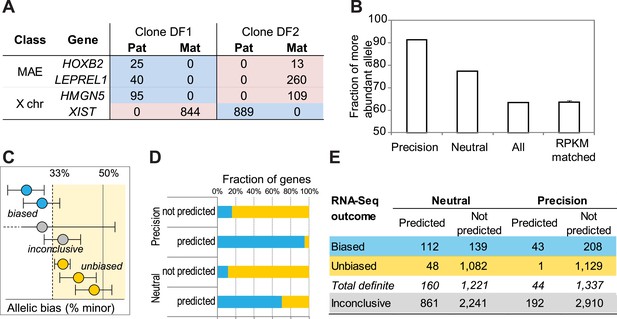
Prediction testing with RNA-Seq.
(A) Representative examples of allelic counts in data from two clones (DF1 and DF2) derived from GM12878 cells. Shown are total maternal (Mat; ‘pink’) or paternal (Pat; ‘blue’) counts for X-linked genes and autosomal monoallelic genes illustrating that the direction of allelic bias is clone-specific. (B) Mean allelic bias in different groups of genes in DF1 and DF2 clones as assessed by the RNA-Seq analysis. ‘50’ corresponds to perfect balance between alleles; ‘100’ to perfectly monoallelic expression. ‘Precision’ and ‘Neutral’—all informative expressed genes predicted as MAE using corresponding settings of the DT2F classifier; ‘All’—all informative expressed genes from GM12878 cells; ‘RPKM matched’—predicted biallelic genes, matched by the expression level to the predicted MAE genes (shown are the mean and standard deviation for 10 permuted sets of genes). (C) Definitions of allelic bias and lack of bias. Unbiased genes (gold) pass equivalence test (with 2:1 boundaries in either direction; equivalence area is light yellow); biased (blue) pass binomial test; genes that pass neither statistical test are called inconclusive (gray). See Figure 2—figure supplement 1 for X-chromosome analysis results according to this scheme. (D) Fraction of genes showing allelic bias in DF1 and DF2 clones as assessed by RNA-seq. Biased genes were called in at least one clone based on FDR-corrected binomial test and displayed at least 2:1 bias. Unbiased genes were called based on passing the equivalence test in at least on clone and not passing the bias test in the other clone. (E) Allele-specific analysis of RNA-Seq data from DF1 and DF2 clones. Experimentally determined allelic states of autosomal genes. Predicted monoallelic and biallelic status is based on the neutral DT2F classifier. Assignments of genes as biased, unbiased or inconclusive (indeterminate in both clones). Color-coding as in panel C.

Allelic bias calling on X-Chromosome.
Application of the bias definition to X-linked genes in the DF1 and DF2 clones. Each rectangle corresponds to one gene; only informative genes are shown; genes are in the correct order on the chromosome but positions are not to scale. Blue—biased with paternal expression; magenta—biased with maternal expression (as defined in Figure 2C).
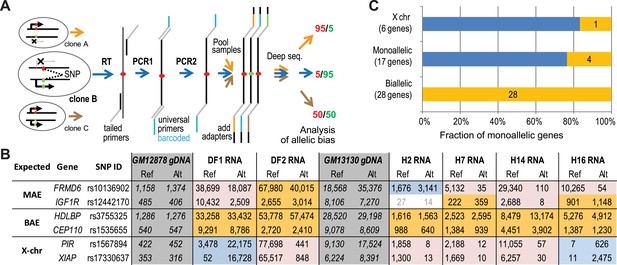
Prediction testing with allele-specific targeted sequencing (AST-Seq).
(A) Schematic representation of the deep barcoding approach. As an illustration, analysis of three clones with no multiplexing is shown, each with a different allelic bias at a SNP of interest. Random-primed cDNA or genomic DNA are used as templates for PCR1, using gene-specific primers with universal tails. The next step associated universal amplicon tails in each sample with two barcodes (PCR2); this allows for barcoding a large number of samples with limited number of secondary primers. For a given sample, all amplicons share the same two barcodes. Barcoded amplicons from all samples are pooled, and sequencing adaptors attached. After sequencing and deconvolving by barcode, allelic hits are counted. (B) Representative allelic counts using the AST-seq. Allelic bias was assessed in two clonal lines, DF1 and DF2, derived from GM12878 and four clones, H2, H7, H14, and H16, from GM13130 (‘H0’) cells. Target SNPs were chosen to be informative in both cell lines. Genomic DNA (gray) was used as a control for allelic bias introduced in amplification; only unbiased assays were pursued. Shown are representative assays for X-linked genes (as control), and examples of genes predicted MAE or biallelic based on the chromatin signature in GM12878. Pink: expression bias towards reference (Ref) allele; blue: expression bias towards alternative (Alt) allele; gold: unbiased expression; no color: counts below threshold—data ignored. Note that, as expected, genes with clone-specific MAE could be biallelic in some clones. (C) Summary of the AST-seq analysis for all tested genes in six clonal samples. Biased (blue) and unbiased (gold) expression as defined in Figure 2C.
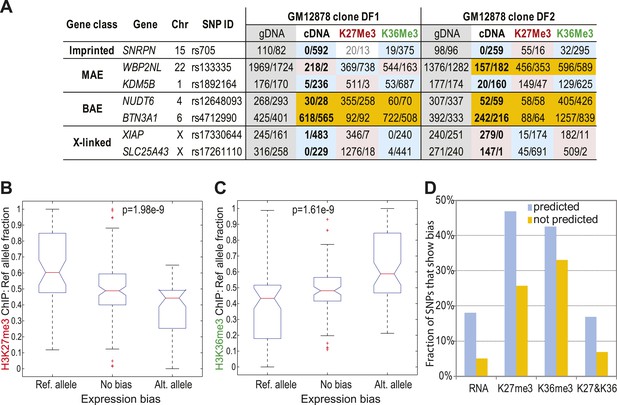
Correlation of allelic bias in expression with bias in chromatin marks.
(A) Representative examples of allelic counts in SNPs assessed with multiplexed targeted sequencing using padlocked probes. Apart from shown clones, additional clones from GM13130 individual were assessed (see ‘text’). Shown as control are an imprinted gene SNRPN and X-linked genes. Other genes were predicted MAE or biallelic based on their gene-body chromatin signature in GM12878 cells. Measurement for each SNP is shown as read counts for the reference and alternative (Ref/Alt) alleles as designated in dbSNP. Color-coding as in Figure 3B. Analysis summarized in this figure is based on 482 SNPs within 458 genes. (B) Correlation of allelic bias in H3K27me3 with allelic bias in cDNA. All informative SNPs were put into one of three bins according to their cDNA allelic bias: unbiased (Figure 2C); significantly biased towards reference allele; significantly biased towards alternative allele. For each of these groups, allelic bias for the ChIP sample from the same clone was assessed and analyzed using Kruskal–Wallis non-parametric ANOVA test. (C) Same as B, for H3K36me3. (D) Fraction of SNPs in predicted MAE and biallelic genes, showing allelic bias of 2:1. Bias calls were made by binomial testing in cDNA, H3K36me3 and H3K27me3 ChIP for SNPs, using data from the padlock probe experiments.
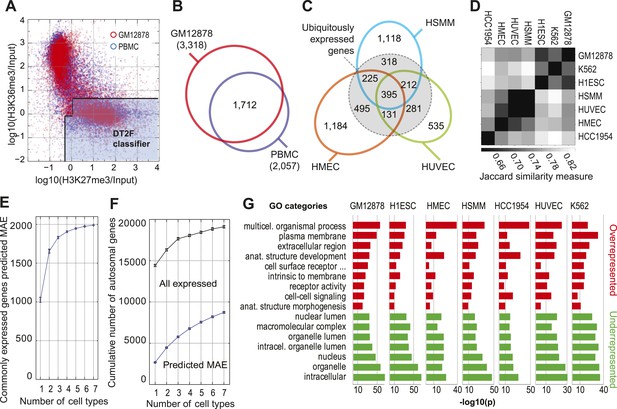
MAE chromatin signature in multiple cell types.
(A) Comparison of H3K27me3 and H3K36me3 ChIP-Seq gene body signal distribution for the autosomal genes in GM12878 cells (red) and in primary peripheral blood monocytes (PBMC; blue). Silent genes (RPKM < 0.1) are excluded in either case. Both datasets were collected by ENCODE; PBMC data: GSE16368. Note that the dots are made more transparent than in Figure 1 to make clear the overall shape of the distribution. (B) Overlap of predicted MAE genes in GM12878 and PBMC (silent genes are excluded). (C) Tissue-specific distribution of MAE genes. Overlap between predicted MAE genes in three cell types as labeled. Within dotted circle: genes expressed in all three lines (and MAE in at least one). Outside dotted circle: MAE genes showing cell type-specific expression (predicted MAE and expressed in that cell type, but silent in at least one of the two other cell lines). (D) Similarity of predicted MAE gene sets in seven cell types: GM12878—lymphoblasts, K562—acute myelocytic leukemia, H1ESC—human embryonic stem cells, HSMM—human skeletal muscle myocytes, HUVEC—human vascular epithelium, HMEC—human mammary epithelium, HCC1954—breast cancer. Similarity assessed according to the Jaccard similarity measure. In the heat map, darker gray corresponds to higher similarity. ChIP-Seq and RNA-Seq data sources: see Dataset S1 and S2 in Dryad (Nag et al., 2013). (E) Cumulative number of predicted MAE genes as a function of the number of cell lines assessed. Counted are only genes expressed in all analyzed cell lines. Order of addition of cell lines was sampled by permutation, shown are mean values ± standard deviation. (F) Cumulative number of all genes and predicted MAE genes as a function of the number of cell lines assessed. Counted are all genes with any evidence of expression in at least one cell type. (G) Gene Ontology (GO) analysis of genes predicted MAE in indicated cell types. Most over- and under-represented categories for GM12878 cells are also shown for other cell types (in each cell line, predicted MAE genes are compared to all expressed genes). Horizontal axis: −log10(p), after Benjamini–Hochberg correction. Gray lines correspond to −log10(p) values as noted.
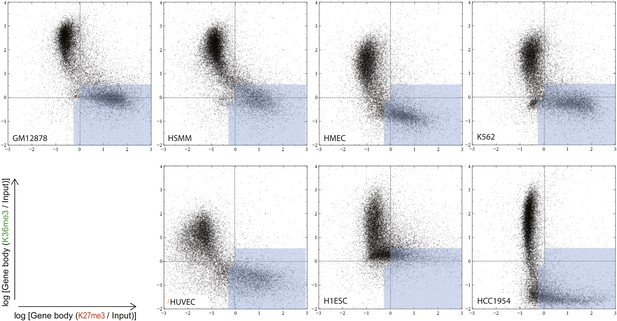
Overall distributions of the normalized H3K27me3 and H3K36me3 gene body signal in the analyzed cell types.
Comparison of H3K27me3 and H3K36me3 ChIP-Seq gene body signal distribution for the autosomal genes in a number of human cell lines (as noted). Each dot represents an autosomal gene. Silent genes (RPKM < 0.1) are excluded. Light blue area illustrates partitioning of this space by the most optimal classifier (DT2F).
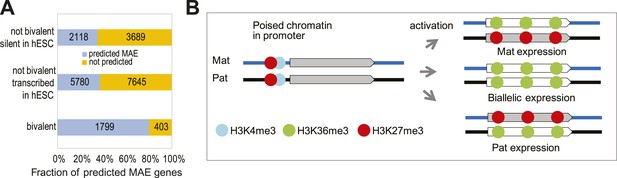
MAE chromatin signature in bivalent genes.
(A) Overrepresentation of bivalent genes among predicted MAE genes. Predicted: predicted MAE in at least one cell line and not silent (RPKM > 0.1); not predicted: not predicted MAE in any cell line. Groups of genes: (top) not bivalent, silent in hESC (RPKM < 0.1); (middle) not bivalent, not silent in hESCs; (bottom) reported bivalent in human ES cells. (B) A speculative model of MAE establishment in bivalent genes. Genes with bivalent/poised chromatin in promoters are silent in undifferentiated stem cells; two alleles have symmetric distribution of active and inactive histone marks. When such gene is activated upon reaching a point of cell fate determination, either one of the alleles (or both) can become transcriptionally active. The initial choice is stochastic, but it is stable in the clonal progeny. Asymmetric histone modifications in the gene body reflect activity of the alleles.





