Evolution of extreme resistance to ionizing radiation via genetic adaptation of DNA repair
Figures
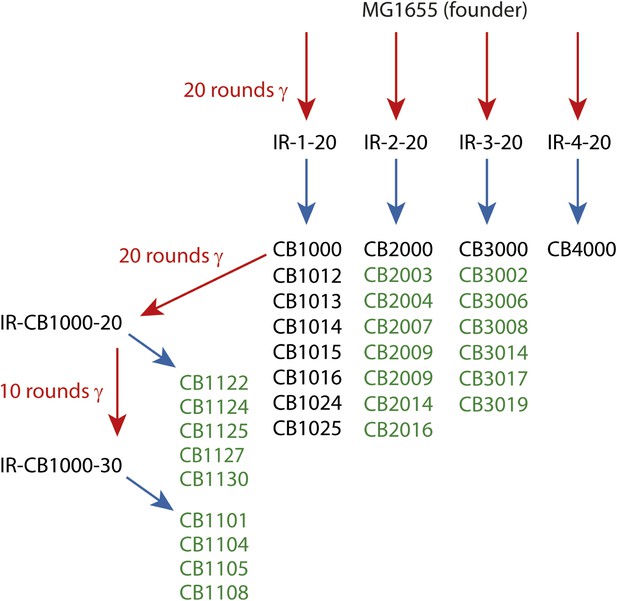
Directed evolution scheme for the evolved isolates described in this paper.
Red arrows denote cycles of irradiation and outgrowth. Evolved populations have titles beginning with “IR”. Isolates were derived from each listed population, as indicated by blue arrows. Isolates from each population are listed under the respective blue arrows, and each isolate features a name beginning with CB. Isolates listed in green text are described for the first time in this study. The sequences of the remaining isolates were described previously (Harris et al., 2009), and are listed here since the genomic data was utilized in the current analysis.
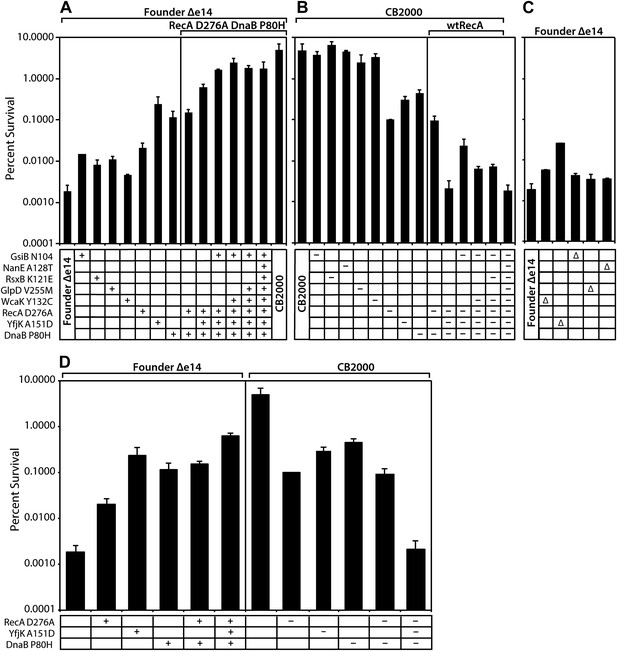
Effects of selected mutations on survival of E. coli.
Mid-logarithmic phase cultures were irradiated to 3000 Gy and plated to measure survival as described in ‘Material and methods’. (A) mutations discovered in CB2000 were moved individually and in combination into the Founder Δe14 background. Mutations present in a given strain are indicated by a + symbol. For reasons not understood, it proved impossible to move the nanE mutation into this background on its own. CB2000 itself is presented in the final lane. (B) the same point mutations (this time including nanE) were mutated back to Founder sequence in the CB2000 background. The first lane is CB2000. Mutations converted to the wild type allele in a given strain are indicated by a–symbol. (C) non-essential genes assayed in Panel A and B were deleted in the Founder Δe14 background, with the deleted gene indicated by a Δ symbol. D, The effects of mutations in genes recA, dnaB, and yfjK are summarized, with symbols as in panels A–C. All results were obtained with a 137Cs irradiator (∼7 Gy/min).
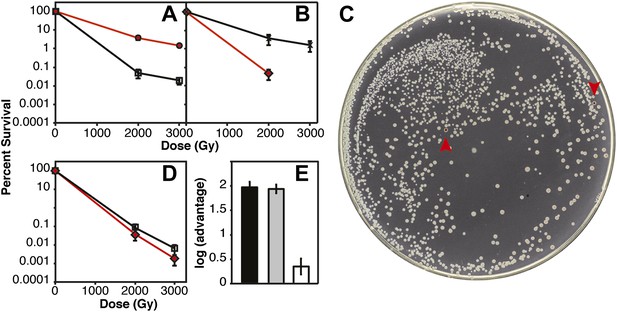
IR survival in direct competition assays.
Mid-logarithmic cultures consisting of a 1:1 ratio of the competiting Ara+ and Ara–strains were irradiated to 2000 and 3000 Gy and plated on tetrazolium arabinose indicator plates to distinguish the frequency of survival for both strains. (A) CB2000 Ara– (red, ^), vs CB2000 wtRecA wtDnaB wtYfjK, ☐ (B) Founder Δe14 Ara−, (red, △), vs Founder Δe14 RecA D276N DnaB P80H YfjK A151D, ◇ (C) TA plate of 2000 Gy survival competition of Founder Δe14 Ara−vs Founder Δe14 RecA D276N DnaB P80H YfjK A151D. The two red Founder Δe14 Ara–colonies are indicated by red arrows. (D) Founder Δe14 Ara− (red, ◇), vs CB2000 wtRecA wtDnaB wtYfjK, ☐. (E) Log advantage in survival to 2000 Gy of CB2000 Ara−over CB2000 wtRecA wtDnaB wt YfjK (in black), Founder Δe14 RecA D276N DnaB P80H YfjK P80H over Founder Δe14 Ara− (in grey), and CB2000 wtRecA wtDnaB wt YfjK over Founder Δe14 Ara− (in white).
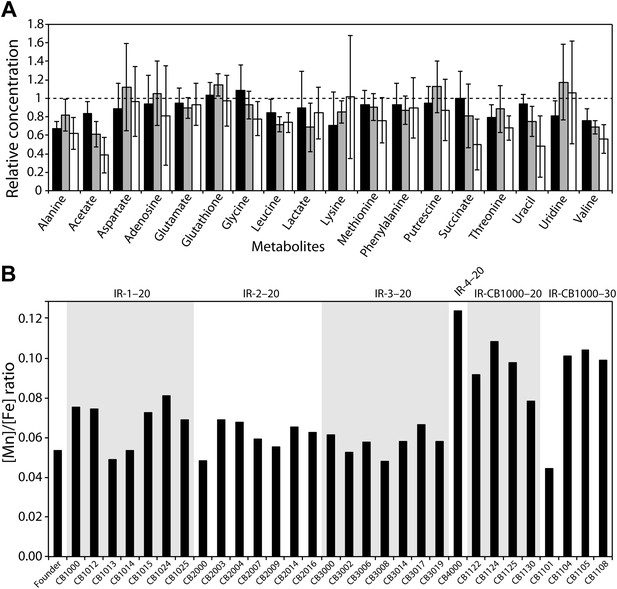
(A), Measurements of metabolites from three representative evolved E. coli strains as compared to Founder.
Metabolites from whole cell pellets collected during logarithmic growth in LB were identified using a two-dimensional 1H-13C Heteronuclear Single Quantum Coherence (HSQC) experiment. Each metabolite is expressed as a ratio of the amount measured in the evolved strain (CB1000, CB1013, or CB2000; black, gray, and white bars, respectively) relative to the Founder. (B) Ratios of manganese to iron are plotted for all isolates for which genomic sequences were obtained. The average increase in Mn/Fe ratio in strains derived from the further evolution of CB1000 is 1.4-fold.
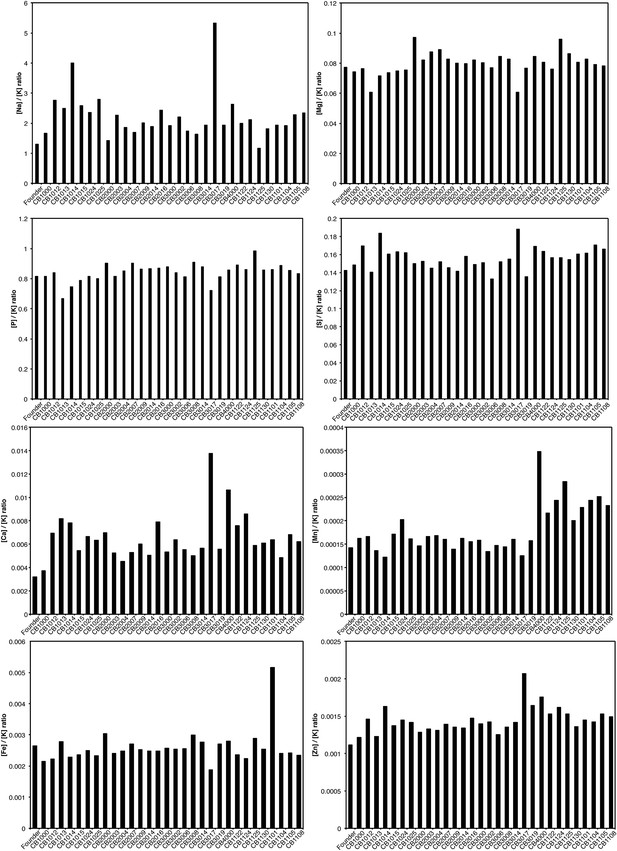
Relative levels of metals are unchanged in evolved E. coli.
Cells sampled during mid-logarithmic growth were subjected to trace metal analysis as described in ‘Materials and methods’. The amount of each metal was plotted as a ratio normalized to the amount of potassium present in each sample, K, which was chosen arbitrarily to correct for variation in metal extraction.
Tables
Summary of the mutational spectrum observed in strains derived from directed evolution of resistance to ionizing radiation
| IR-1-20 | IR-2-20 | IR-3-20 | IR-4-20 | IR-CB1000-20 | IR-CB1000-30 | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transitions | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C |
| B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | |
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| 0 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 2 | 2 | 2 | 2 | 3 | 0 | 0 | 0 | 0 | |
| 0 | 2 | 3 | 4 | 5 | 4 | 5 | 0 | 3 | 4 | 7 | 9 | 4 | 6 | 0 | 2 | 6 | 8 | 4 | 7 | 9 | 0 | 2 | 4 | 5 | 7 | 0 | 1 | 4 | 5 | 8 | |
| C → T | 19 | 20 | 6 | 10 | 15 | 6 | 9 | 12 | 13 | 11 | 12 | 11 | 9 | 9 | 14 | 14 | 13 | 14 | 13 | 12 | 12 | 14 | 59 | 64 | 69 | 69 | 63 | 60 | 62 | 63 | 62 |
| G → A | 18 | 9 | 11 | 16 | 11 | 11 | 12 | 17 | 17 | 16 | 15 | 13 | 13 | 11 | 17 | 11 | 13 | 13 | 12 | 8 | 15 | 10 | 60 | 65 | 78 | 71 | 60 | 57 | 60 | 60 | 60 |
| T → C | 7 | 7 | 6 | 4 | 6 | 13 | 4 | 11 | 8 | 16 | 10 | 9 | 9 | 11 | 11 | 4 | 13 | 11 | 7 | 6 | 9 | 7 | 51 | 51 | 40 | 44 | 47 | 47 | 47 | 47 | 47 |
| A → G | 15 | 16 | 7 | 8 | 8 | 14 | 11 | 10 | 6 | 6 | 7 | 7 | 6 | 5 | 6 | 14 | 10 | 10 | 9 | 11 | 6 | 8 | 42 | 43 | 42 | 54 | 50 | 46 | 50 | 50 | 50 |
| Transversions | |||||||||||||||||||||||||||||||
| G → T | 4 | 4 | 1 | 1 | 1 | 3 | 5 | 3 | 4 | 2 | 3 | 5 | 4 | 2 | 5 | 5 | 4 | 4 | 4 | 6 | 4 | 2 | 10 | 9 | 7 | 8 | 6 | 5 | 6 | 6 | 6 |
| C → A | 1 | 1 | 2 | 1 | 1 | 5 | 6 | 5 | 3 | 6 | 3 | 3 | 4 | 6 | 2 | 4 | 2 | 1 | 3 | 4 | 1 | 6 | 3 | 2 | 5 | 5 | 6 | 4 | 5 | 5 | 4 |
| A → C | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 7 | 4 | 2 | 2 | 4 | 4 | 3 | 3 | 3 | |||||||||
| T → G | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 2 | 4 | 1 | 1 | 1 | 1 | 1 | ||||||||||
| G → C | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 2 | 4 | 3 | 3 | 3 | 3 | ||||||||||
| C → G | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | ||||||||||||
| A → T | 1 | 2 | 1 | 1 | 3 | 1 | 2 | 1 | 4 | 3 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 2 | 1 | 2 | 3 | 3 | 3 | 3 | 3 | ||||||
| T → A | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 2 | 1 | 3 | 3 | 2 | 3 | 3 | 3 | 3 | 3 | 2 | ||||||
| Insertions | |||||||||||||||||||||||||||||||
| 767bp IS1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||||||||||||
| Deletions | 1 | ||||||||||||||||||||||||||||||
| e14 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| 93bp | 1 | ||||||||||||||||||||||||||||||
| Totals | 70 | 63 | 40 | 46 | 49 | 56 | 53 | 66 | 55 | 64 | 56 | 53 | 50 | 51 | 63 | 57 | 63 | 61 | 55 | 53 | 54 | 53 | 243 | 251 | 249 | 265 | 250 | 236 | 246 | 247 | 244 |
-
The mutational spectrum of the population is inferred based on the genetic alterations observed in the subset of strains sequenced.
Summary of prominent mutational patterns observed in multiple evolved populations
| IR-1-20 | IR-2-20 | IR-3-20 | IR-4-20 | IR CB1000−+ | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Position | Ref. | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | C | Change | Mutation | |
| B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | B | ||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 4 | ||||||
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| 0 | 1 | 1 | 1 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | ||||||
| Allele | 0 | 2 | 3 | 4 | 5 | 4 | 5 | 0 | 3 | 4 | 7 | 9 | 4 | 6 | 0 | 2 | 6 | 8 | 4 | 7 | 9 | 0 | Type | ||||
| clpP | 456127 | A | G | G | + | Y75C | N | ||||||||||||||||||||
| clpP/clpX | 456637 | G | A | A | A | A | A | A | A | - | I | ||||||||||||||||
| clpX | 457803 | A | G | Y384C | N | ||||||||||||||||||||||
| gsiB | 868947 | A | G | G | G | G | G | G | G | N104S | N | ||||||||||||||||
| gsiB | 869499 | T | C | C | C | C | C | C | C | L288P | N | ||||||||||||||||
| gsiB | 870075 | T | C | V480A | N | ||||||||||||||||||||||
| fnr | 1396995 | A | T | T | T | T | T | T | T | + | F185I | N | |||||||||||||||
| rsxB | 1704735 | A | G | G | G | G | G | G | G | + | K121E | N | |||||||||||||||
| rsxD | 1707299 | T | C | C | C | C | C | C | C | V44A | N | ||||||||||||||||
| wcaM | 2113451 | T | C | + | N156S | N | |||||||||||||||||||||
| wcaK | 2116031 | T | C | C | C | C | C | Y132C | N | ||||||||||||||||||
| wcaC | 2129153 | A | G | G | G | G | G | G | G | S313S | S | ||||||||||||||||
| yfjK | 2759609 | G | A | H651Y | N | ||||||||||||||||||||||
| yfjK | 2760683 | T | C | K293E | N | ||||||||||||||||||||||
| yfjK | 2760809 | G | A | P251S | N | ||||||||||||||||||||||
| yfjK | 2761108 | G | T | T | T | T | T | T | T | A151D | N | ||||||||||||||||
| recA | 2820924 | C | A | A289S | N | ||||||||||||||||||||||
| recA | 2820962 | T | G | G | G | G | D276A | N | |||||||||||||||||||
| recA | 2820963 | C | T | T | T | T | T | T | T | T | D276N | N | |||||||||||||||
| nanE | 3368674 | C | T | T | T | T | T | T | T | + | A128T | N | |||||||||||||||
| nanT | 3369380 | A | G | G | G | G | G | G | G | F405S | N | ||||||||||||||||
| dnaB | 4262560 | T | C | + | L74S | N | |||||||||||||||||||||
| dnaB | 4262578 | C | A | A | A | A | A | A | A | P80H | N | ||||||||||||||||
| dnaB | 4262935 | C | A | P199Q | N | ||||||||||||||||||||||
| priA | 4123174 | C | T | T | T | T | T | T | T | V553I | N | ||||||||||||||||
| priC | 489549 | A | G | G | L162P | N | |||||||||||||||||||||
| dnaT | 4599105 | G | A | A | A | A | R145C | N | |||||||||||||||||||
-
Entries in red denote mutations that are present in CB2000.
Additional files
-
Supplementary file 1
(A) Mutation table for isolates sequenced from IR-1-20, IR-2-20, IR-3-20, and IR-4-20. (B) Mutation table for isolates sequenced from IR-CB1000-20 and IR-CB1000-30. (C) RNA-SEQ results for CB1000 and CB2000 as compared to Founder. (D) Strains used in this study.
- https://doi.org/10.7554/eLife.01322.010





