tRNA genes rapidly change in evolution to meet novel translational demands
Figures
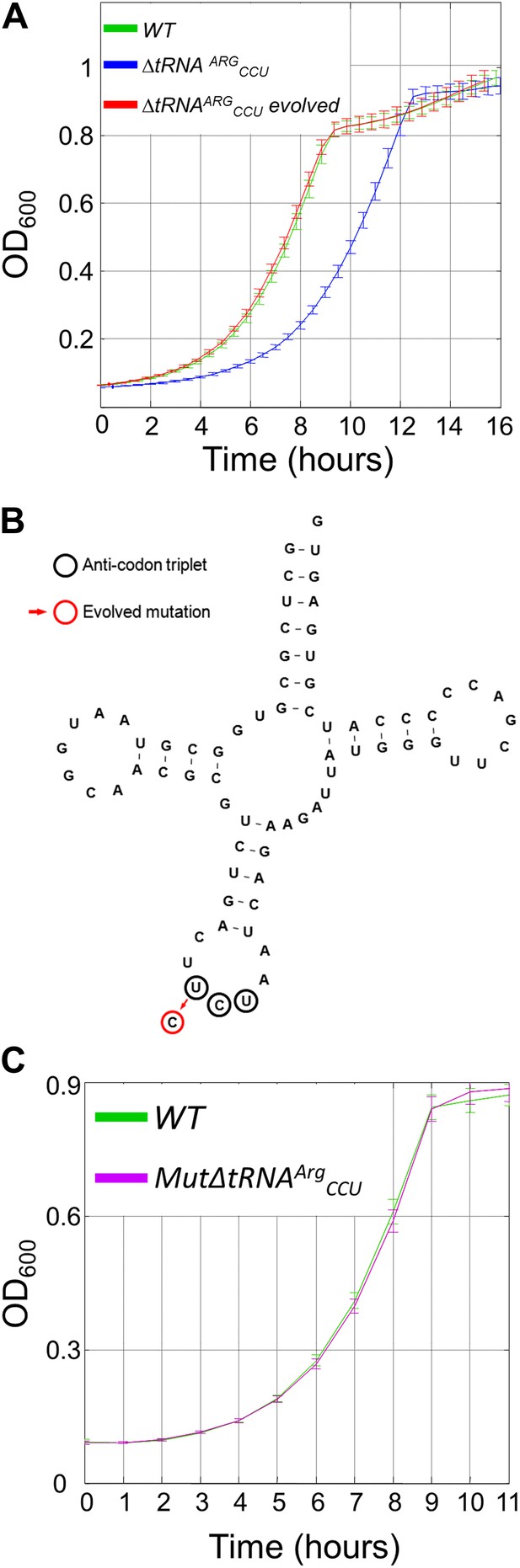
The growth defect associated with deletion of a singleton tRNA gene was rapidly rescued during the lab-evolution experiment.
(A) Growth curve measurements of wild-type (WT) (green), ΔtRNAArgCCU (blue) and the evolved deletion (red) are shown in optical density (OD) values over time during continuous growth on rich medium at 30°C. (B) The mutation that was found to recover the deletion phenotype in the evolved strains is shown on the secondary structure of tRNAArgUCU. The UCU anticodon nucleotides are marked with black circles, and the red circle indicates the mutation that occurred in the anticodon, that is T→C transition. (C) MutΔtRNAArgCCU in which the same mutation that was found in the evolved strain was deliberately engineered, exhibits similar growth as the WT. Growth curve measurements of WT (green) and of MutΔtRNAArgCCU (magenta) are shown in OD600 values over time during continuous growth on rich medium at 30°C.
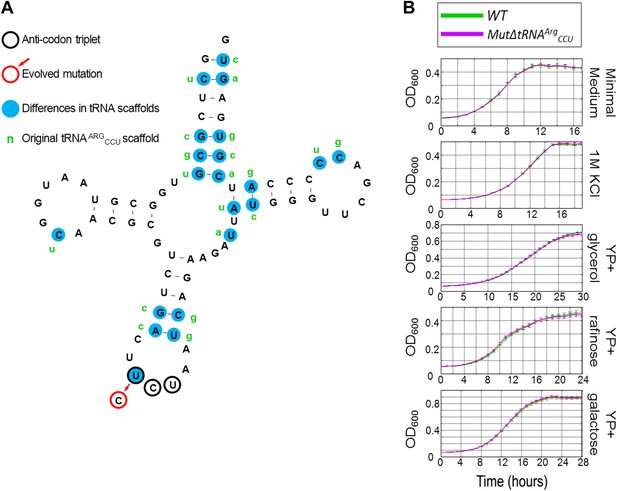
The growth of MutΔtRNAArgCCU carrying the chimeric tRNA compared to wild-type (WT) under different conditions.
(A) The sequence of the chimeric tRNA is drawn showing the scaffold of tRNAArgUCU with the mutated CCU anticodon. The anticodon triplet is marked with black circles. The evolved mutation is marked with a red circle. All 20 nucleotide differences between tRNAArgUCU and tRNAArgCCU are marked with blue background, next to which, in green letters, the original nucleotide of tRNAArgCCU are written. (B) Growth curve measurements of WT (green) and of MutΔtRNAArgCCU (magenta) are shown in OD600 values over time during continuous growth.
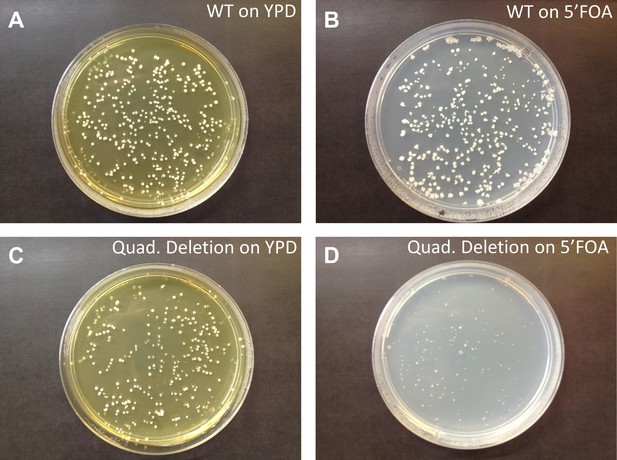
Quadruple deletion of tRNAserGCU is lethal.
We perturbed the tRNA pool in a wild-type strain by deletion of an entire serine tRNA family. Here, the supply of tRNASerGCU was eliminated by deletion of all four identical gene copies of this family located on chromosomes IV, VI, XII and X. A complete deletion of this gene family is lethal, indicating that the tRNASerGCU is essential in Saccharomyces cerevisiae. To validate that the lethality is indeed due to the deletion of the tRNASerGCU genes and not due to an unintentional perturbation of other putative genetic features in the vicinity of the deleted tRNASerGCU copies, we introduced a plasmid with the tRNASerGCU gene. As expected, the quadruple deletion strain was viable when supplemented with a plasmid carrying the tRNASerGCU gene. (A and C) BY384 and tRNASerGCU quadruple deletion strains, respectively, with a plasmid harboring a tRNASerGCU gene grown on yeast extract/peptone/dextrose (YPD) plates. (B and D) BY384 and tRNASerGCU quadruple deletion strains, respectively, grown on liquid medium that contains uracil (YPD) to allow the growth of cells that lost the plasmid and then plated on 5-fluoro-orotic acid (5-FOA) plates (see ‘Materials and methods’). As expected, normal size colonies were only observed in A, B and C. This means that the quadruple deletion strain with the plasmid could not lose the tRNA gene because of its essentiality.
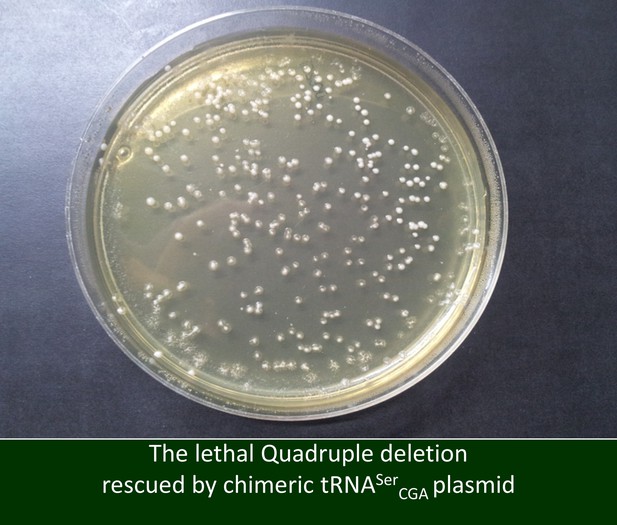
A chimeric serine tRNA can rescue the lethality of the quadruple deletion.
Here, rather than inserting a plasmid with the original tRNASerGCU gene, we complemented the tRNASerGCU family deletion strain with a plasmid containing a chimeric serine tRNA with a GCU anticodon. The chimeric tRNA has an alternative scaffold derived from a different serine tRNA, tRNASerCGA, that differs in 22 positions from the deleted serine tRNA family. This strain formed normal size colonies on a yeast extract/peptone/dextrose (YPD) plate.
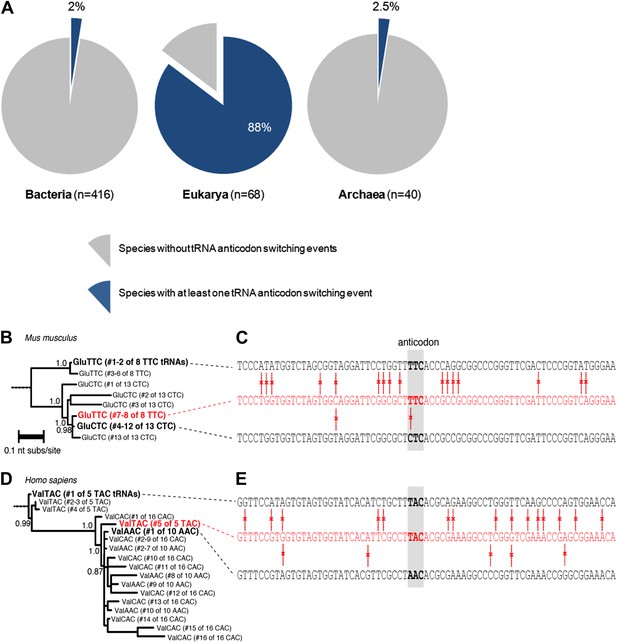
Anticodon switching is a widespread phenomenon in nature.
(A) Number of species with at least one tRNA switching event in each domain of life. (B) The anticodon UUC convergently evolved in Mus musculus. A maximum likelihood phylogeny of tRNA sequences in M. musculus that decode glutamic acid (Glu) codons. Branch lengths express average nucleotide substitutions per site. Decimals on internal branches express branch support. (C) A comparison of nucleotide sequences for glutamic acid tRNA genes in M. musculus with anticodon UUC (top, tRNA1547 and tRNA359), ‘switched’ UUC tRNAs (middle, tRNA286 and tRNA754), and CUC tRNAs (bottom, tRNA1002, tRNA745, tRNA303, tRNA999, tRNA996 tRNA709, tRNA1001, tRNA1912 and tRNA81). The anticodon triplet is boxed in gray. Red vertical bars indicate differences between sequences. (D) The anticodon UAC convergently evolved in Homo sapiens. A maximum likelihood phylogeny of tRNA sequences in H. sapiens encoding for valine (Val) is shown. (E) A comparison of nucleotide sequences for H. sapiens tRNAs with anticodons UAC (top, tRNA6), a ‘switched’ UAC tRNA (middle, tRNA40), and an AAC tRNA (bottom, tRNA136). The number of genes is according to the tRNA database.
-
Figure 3—source data 1
Table of anticodon switchings in different species across the tree of life.
- https://doi.org/10.7554/eLife.01339.008
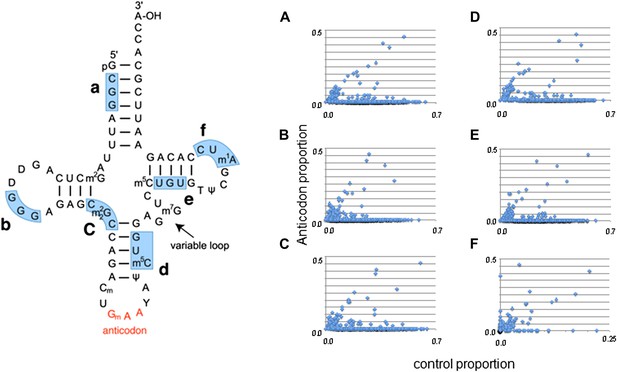
A comparison of discrepancy proportions at the anticodon triplet vs control triplets.
Six control triplets were chosen; their locations are marked on a tRNA structure. (A–F) A comparison of the proportion of switched tRNAs for each species when the anticodon was masked versus the proportion of alternative discrepancies when the control triplet was masked. Each point corresponds to one species. A tRNA was identified as ‘switched’ if its nucleotide sequence phylogenetically clustered with other tRNAs with dissimilar mask triplets (see ‘Materials and methods’).
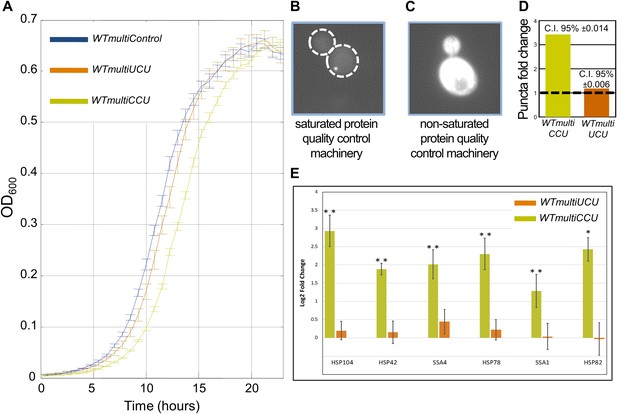
WTmultiCCU experiences a growth defect compared to WTmultiUCU and demonstrates higher levels of misfolded proteins.
(A) Growth curve measurements of WTmultiControl (blue), WTmultiUCU (brown) and WTmultiCCU (khaki) are shown in optical density (OD) values over time during continuous growth. The WTmultiCCU strain carrying a high copy number plasmid harboring tRNAArgCCU demonstrates slower growth compared to cells with an empty plasmid or with a tRNAArgUCU plasmid that is mainly characterized by a longer growth delay (lag phase). (B) A demonstration of a WTmultiCCU cell in which the mCherry-Von Hippel–Lindau (VHL) proteins appear with a punctum phenotype when the protein quality control machinery is saturated with misfolded proteins. (C) A demonstration WTmultiUCU cell in which the quality control machinery is not occupied with other proteins; mCherry-VHL is localized to the cytosol. (D) WTmultiCCU, WTmultiUCU and WTmultiControl were transformed with a VHL-mCherry containing plasmid and visualized under the microscope; 1000 cells per strain were counted for either cytosolic or punctum localization of the VHL protein. The fold change in the number of cells containing puncta was then deduced by normalization to the WTmultiControl population. The 95% confidence interval is indicated. (E) The mRNA fold change of six representative heat-shock genes measured by real-time quantitative PCR (RT-qPCR). Presented values are the mean of two biological repetitions ± SEM. The significance of the fold change differences was examined using a t test, with *p<0.001 or **p<0.0001.
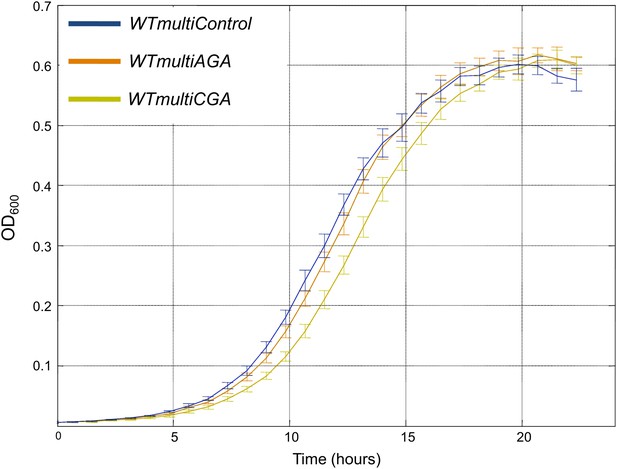
Multiple copies of rare tRNASerCGA gene are deleterious compared to abundant tRNASerAGA.
Growth curve measurements of WTmultiControl (blue), WTmultiAGA (brown) and WTmultiCGA (khaki) are shown in optical density (OD) values over time during continuous growth on rich medium at 30°C. The WTmultiCGA strain carrying a high copy number plasmid harboring tRNASerCGA demonstrates slower growth compared to cells with an empty plasmid or with a tRNASerAGA plasmid.
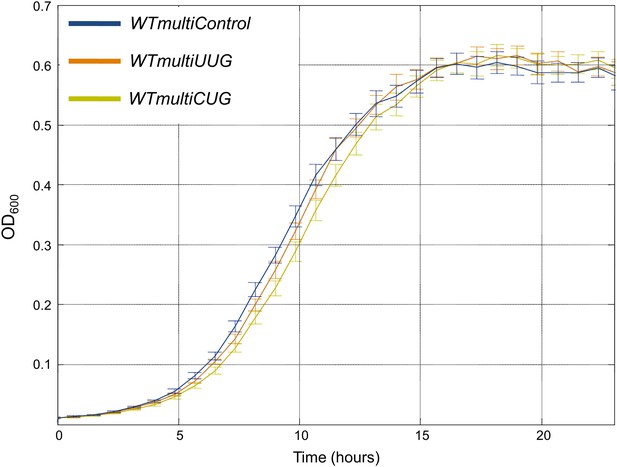
Multiple copies of the rare tRNAGlnCUG gene are deleterious compared to abundant tRNAGlnUUG.
Growth curve measurements of WTmultiControl (blue), WTmultiUUG (brown) and WTmultiCUG (khaki) are shown in optical density (OD) values over time during continuous growth on rich medium at 30°C. The WTmultiCUG strain carrying a high copy number plasmid harboring tRNAGlnCUG demonstrates slower growth compared to cells with an empty plasmid or with a tRNAGlnUUG plasmid.
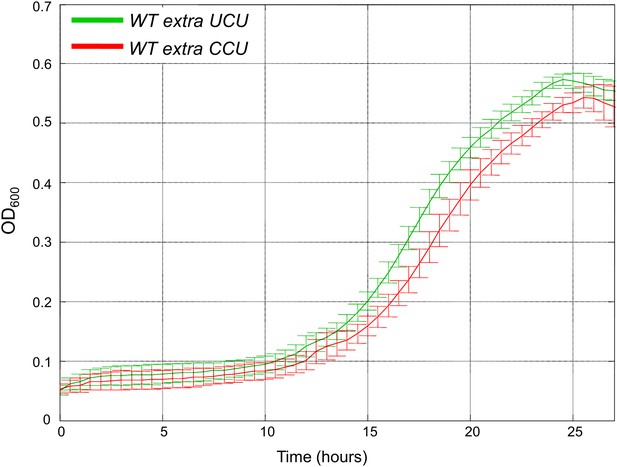
Addition of low copy number tRNAArgCCU is deleterious compared to low copy number tRNAArgUCU when grown in heat.
We created two strains harboring low copy number (CEN) plasmids carrying either tRNAArgUCU or tRNAArgCCU in addition to the endogenous copy. We termed these strains WTextraUCU and WTextraCCU, respectively. Growth curve measurements of WTextraUCU (green) and WTextraCCU (red) are shown in optical density (OD) values over time during continuous growth on rich medium at 39°C. Cells with the centromeric tRNAArgCCU plasmid showed a modest growth defect compared to cells with the centromeric tRNAArgUCU plasmid.
Additional files
-
Supplementary file 1
Usage of arginine codons in Saccharomyces cerevisiae.
(A) The AGG codon constitutes approximately 21% of the arginine codons in the yeast genome and approximately 16% of the arginine codons in the yeast transcriptome under standard lab conditions. In comparison, the AGA codon constitutes approximately 47.5% of the arginine codons in the genome and approximately 56% of the arginine codons in the transcriptome (Gingold et al., 2012). (B) Statistical significance of differences between proportions of discrepancies when masking the anticodon triplet versus control triplets. (a) Six control triplets were masked (Figure 3—figure supplement 1), and the analysis to identify switched tRNAs was repeated (see ‘Materials and methods’). For each masked triplet, we computed the percentage of tRNA sequences that are not clustered according to their triplet content in each species. (b) The proportion of switched tRNAs was averaged across all species, and (c) the standard error of this average was computed. (d) The distribution of tRNA switching proportions from the original anticodon analysis was compared to the distribution of discrepancies from the control triplet, using a paired t test. The p value from this comparison is reported here. (C) Various yeast species tend to keep tRNAArgCCU in a single copy. All examined yeast species maintain a single copy of tRNAArgCCU compared to tRNAArgUCU, which is mostly found in multiple copies. The copy number of tRNAArgUCU and tRNAArgCCU genes is shown together with the codon usage of AGG and AGA codons in the different yeast genomes.
- https://doi.org/10.7554/eLife.01339.014
-
Supplementary file 2
Plasmids and primers.
- https://doi.org/10.7554/eLife.01339.015





