Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling
Figures
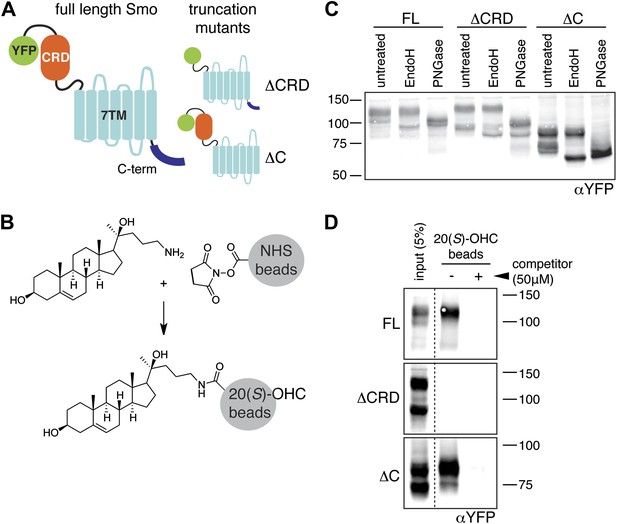
The mouse Smo CRD is required to bind oxysterols.
(A) Schematic of full-length (FL), YFP-tagged mSmo and the ΔCRD and ΔC truncation mutants used in this study. (B) Structure of the 20(S)-OHC beads used in Smo pull-down assays. (C) EndoH and PGNaseF sensitivity of YFP-mSmo, ΔCRD-YFP-mSmo and ΔC-YFP-mSmo stably expressed in Smo−/− cells and loaded on an 8% Tris-Glycine SDS-PAGE gel. The fraction of each protein with slower mobility on the gel was resistant to EndoH but sensitive to PGNaseF, suggesting post-Golgi localization. (D) 20(S)-OHC beads captured YFP-mSmo and ΔC-YFP-mSmo, but not ΔCRD-YFP-mSmo from lysates of cells stably expressing each protein. Binding to beads was not seen when 50 μM free 20(S)-OHC was added as a competitor.
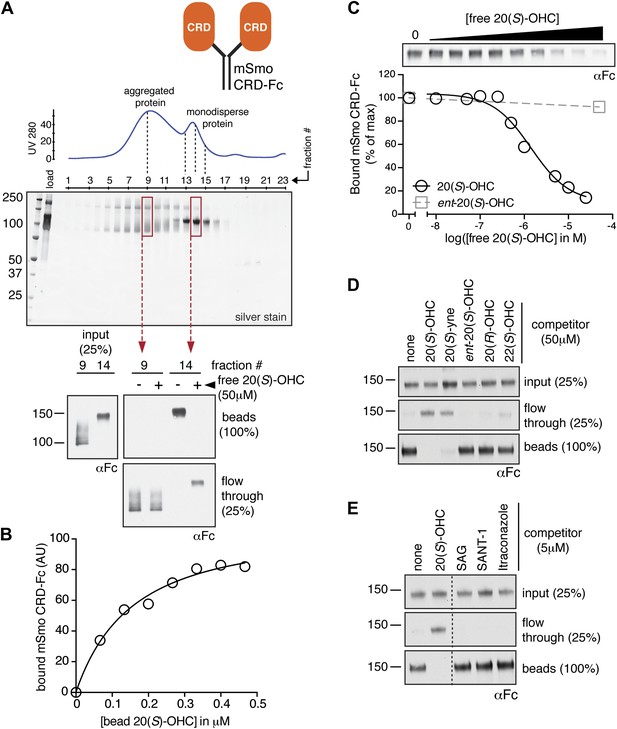
The isolated mSmo CRD can bind oxysterols.
(A) Fractionation of the mSmo CRD-Fc protein on a Superose 6 gel-filtration column. The UV280 absorbance of each fraction (blue curve) is shown above the protein content of each fraction on a silver stained gel. Monodisperse protein (fractions 13–15) elutes in a sharp peak and binds to 20(S)-OHC beads (panels below), while aggregated protein runs as a broad peak (fractions 5–12) and fails to bind oxysterols. The indicated fractions (red boxes) were incubated with 20(S)-OHC beads in the presence or absence of free 20(S)-OHC competitor, and the amount of mSmo CRD-Fc protein captured on the beads or left in the flow through was assayed on an anti-Fc immunoblot. (B) A binding curve (Kd ∼180 nM) for the mSmo CRD-Fc-20(S)-OHC interaction was measured by incubating a fixed amount of protein with increasing amounts of bead-immobilized sterol. (C) Binding of mSmo CRD-Fc to 20(S)-OHC beads is inhibited in a dose-responsive fashion by free 20(S)-OHC but not by the enantiomer ent-20(S)-OHC. A competition assay was used to test the ability of various oxysterols (D) or Smo ligands (E) to inhibit the binding of mSmo CRD-Fc to 20(S)-OHC beads. Anti-Fc immunoblots show the amount of protein in the input, captured on the beads, and left in the flow-through.
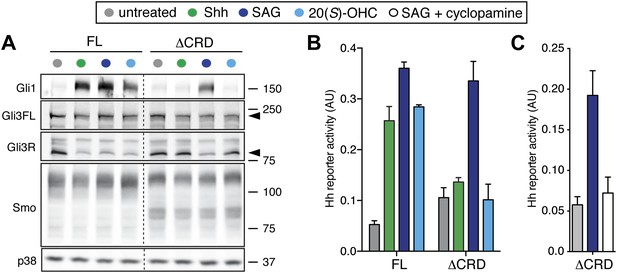
The mSmo CRD is required for Shh- and oxysterol-mediated activation of Hh signaling.
(A) Smo−/− cells stably expressing full-length (FL) YFP-mSmo or ΔCRD-YFP-mSmo were treated with Shh, SAG (100 nM) or 20(S)-OHC (10 μM). Levels of Gli1 and Gli3R protein, determined by immunoblotting after fractionation on an 8% Tris-glycine SDS-PAGE gel, were taken as a metric of pathway activation. An anti-YFP blot shows the levels of YFP-mSmo in each sample, and p38 levels are used as a loading control. (B and C) A luciferase-based Hh reporter gene was used to measure signaling in Smo−/− cells transiently transfected with constructs encoding YFP-mSmo or ΔCRD-YFP-mSmo and then treated with the indicated Smo ligands. In (C), ΔCRD-YFP-mSmo activated with SAG (25 nM) can be inhibited by the co-administration of cyclopamine (5 μM). Error bars denote S.D. (n = 3).
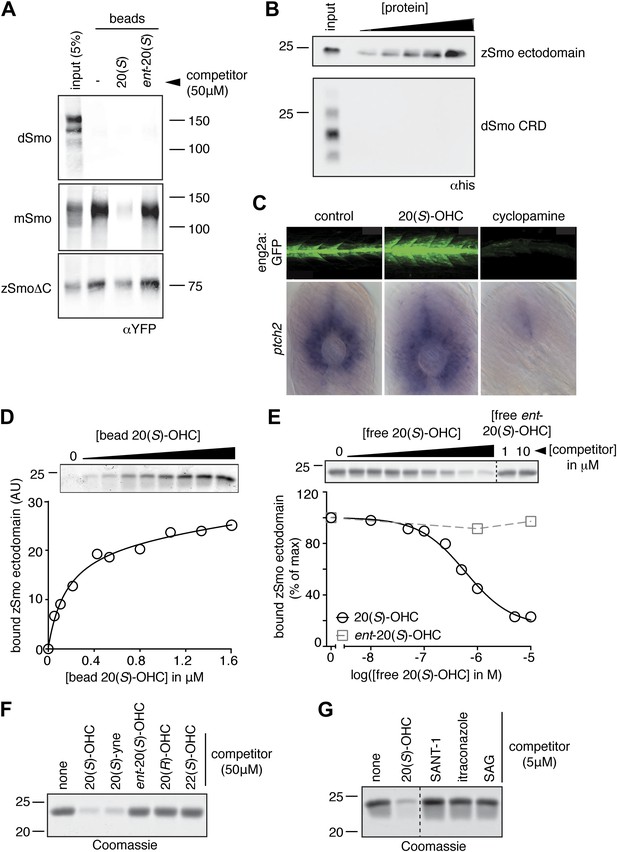
The Smo-oxysterol interaction is conserved in vertebrates.
(A) The interaction of 20(S)-OHC beads with full-length mSmo, full-length Drosophila Smo (dSmo) or zebrafish Smo (zSmo) carrying a truncation of the intracellular C-terminal tail (zSmoΔC) was tested in the presence of free 20(S)-OHC or its enantiomer. (B) The zSmo ectodomain (which includes the CRD) can bind to 20(S)-OHC beads, but the dSmo CRD cannot. (C) Zebrafish embryos (30hpf) carrying a GFP transgene driven by the engrailed2a promoter were treated with 20(S)-OHC (50 µM) or cyclopamine (40 µM) and assessed for GFP expression by fluorescence and ptch2 expression by in situ hybridization. See Figure 4—figure supplement 1 for quantitation. (D) A binding curve (Kd ∼170 nM) for the zSmo ectodomain-20(S)-OHC interaction was measured by incubating a fixed amount of protein with increasing amounts of bead-immobilized sterol. The amount of zSmo ectodomain captured on the beads (shown in the graph) was quantitated from a coomassie-stained SDS-PAGE gel shown above. (E) Binding of the zSmo ectodomain to 20(S)-OHC beads was inhibited in a dose-responsive fashion by free 20(S)-OHC but not by its enantiomer. (F and G) Coomassie-stained SDS-PAGE gels show the amount of zSmo ectodomain captured on 20(S)-OHC beads in the presence of various oxysterols (F) or Smo ligands (G).
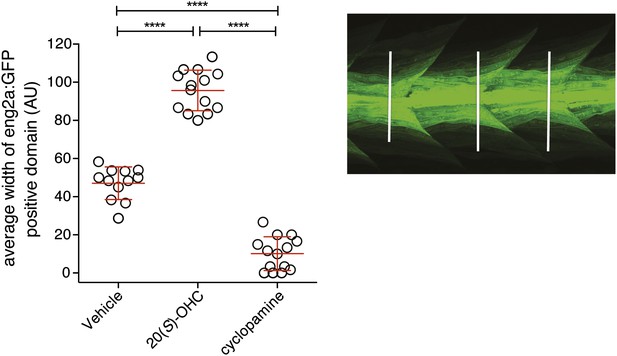
20(S)-OHC activates Hedgehog signaling in zebrafish embryos.
Zebrafish embryos (30hpf) carrying a GFP transgene driven by the engrailed2a (eng2a:GFP) promoter were treated with 20(S)-OHC (50 µM) or cyclopamine (40 µM) and assessed for GFP expression by fluorescence. To quantify the effect of each treatment on eng2a:GFP expression, the width of the GFP-positive domain was measured at three points along the length of each embryo (right panel, white lines) and averaged. The average width is plotted as a scatter plot, with each point representing one embryo. 12–16 embryos per condition are depicted. Red bars represent mean ± SD. All conditions were significantly different from each other (****p<0.0001, one-way ANOVA with Bonferroni correction for multiple comparisons.)
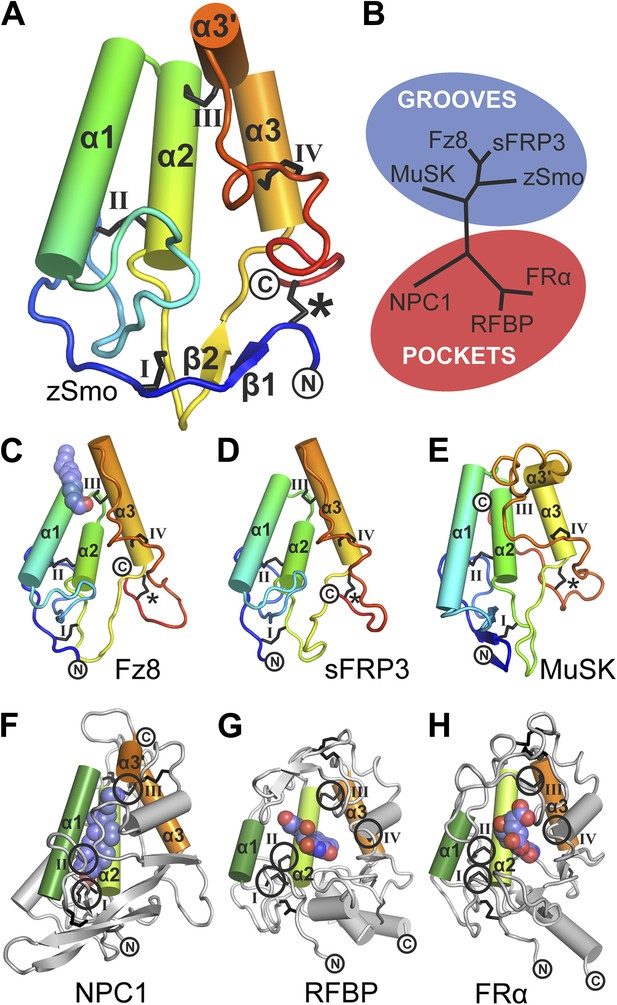
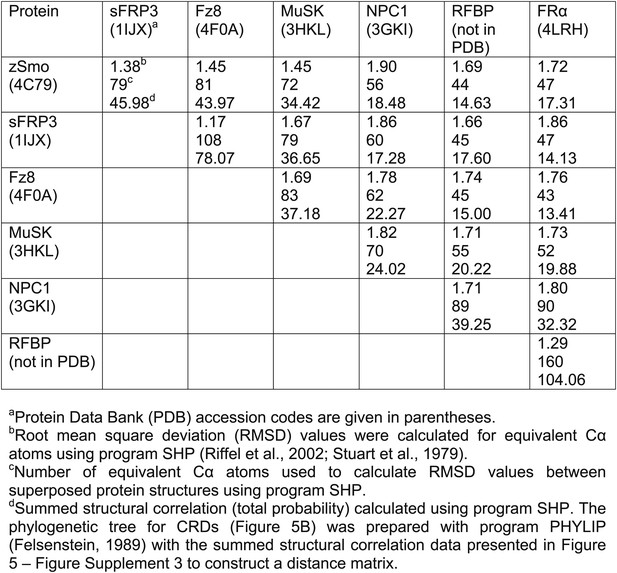
Structural analysis of the zebrafish Smo CRD.
(A) Ribbon diagram of zSmo CRD in rainbow coloring from blue (N-terminus) to red (C-terminus) with the secondary structure elements numbered. The four disulfide bridges (black sticks) conserved in all Fz-like CRDs are depicted with Roman numerals, and the non-conserved disulfide bridge is marked with an asterisk (*). N- and C-termini are labeled. (B) Structural phylogenetic analysis of the CRDs. Structural superposition of CRDs from zSmo, Frizzled 8 (Fz8, PDB ID 4F0A, Janda et al., 2012), secreted Frizzled-related protein 3 (sFRP3, PDB ID 1IJX, Dann et al., 2001), muscle-specific kinase (MuSK, PDB ID 3HKL, Stiegler et al., 2009), Niemann-Pick C1 protein (NPC1, PDB ID 3GKI, Kwon et al., 2009), riboflavin-binding protein (RFBP, Monaco, 1997), and folate receptor α (FRα, PDB ID 4LRH, Chen et al., 2013) were superimposed using SHP (Stuart et al., 1979; Riffel et al., 2002). CRDs that form ligand-binding pockets (red background) or grooves (blue background) form two distinct evolutionary branches. In addition, CRDs show distant structural similarity to the extracellular domains of glypicans (Pei and Grishin, 2012). However, analysis of the crystal structures of glypicans Dally-like protein and glypican 1 revealed no apparent grooves or pockets that could accommodate small molecules (Kim et al., 2011; Svensson et al., 2012) and thus were not included in our structural analyses. (C–H) Ribbon diagrams of superimposed Fz-like CRD domains from the structural phylogenetic analysis in (B). (C) Fz8-palmitoleyl complex, (D) sFRP3, (E) MuSK, (F) NPC1-cholesterol complex, (G) RFBP-riboflavin complex, (H) FRα-folate complex. Color coding and labeling follows (A). Ligands are shown as spheres in atomic coloring (carbon: slate; oxygen: red; nitrogen: blue). In (F–H) the conserved disulfide bridges are highlighted with a circle. NPC1 (F) does not contain disufide bridge IV.
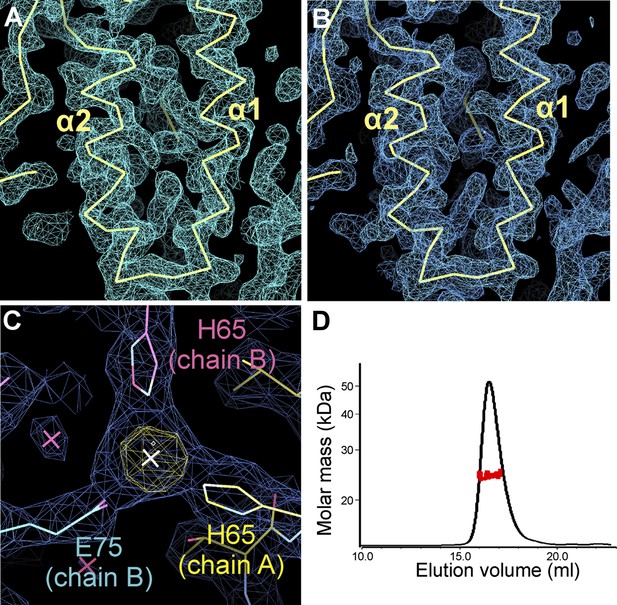
Electron density of the zSmo CRD structure and oligomeric state of the zSmo ectodomain.
(A) SeMet SAD-phased and density modified map from RESOLVE (Terwilliger, 2003) calculated to 2.3 Å resolution and contoured at 1.0 σ showing the two core zSmo CRD helices α1 and α2. (B) SigmaA-weighted 2FO-FC map of the final model of SeMet-labeled zSmo ectodomain from REFMAC (Murshudov et al., 1997) at 2.3 Å resolution and contoured at 1.0 σ. View is the same as in (A). (C) Close-up view of the zinc-binding site in the zSmo CRD crystal structure. The anomalous difference Fourier map (yellow, contoured at 5 σ) and SigmaA-weighted 2FO-FC map (blue, contoured at 1.0 σ) of the final model of native zSmo CRD were calculated to 2.6 Å. Note that zinc is present in a crystal contact formed by three different zSmo chains. (D) Multi angle light scattering of the glycosylated zSmo ectodomain (expressed in mammalian cells) indicates a molecular mass (red scattered dots) of 24.43 ± 0.9 kDa and is in agreement with the theoretical molecular mass for a non-glycosylated monomer (20.4 kDa). The zSmo ectodomain has two predicted N-linked glycosylation sites (each accounting for 2 kDa), which explains the difference between the theoretical and MALS-derived molecular mass. Protein concentration at the elution peak was 8.123×10−5 g/ml.
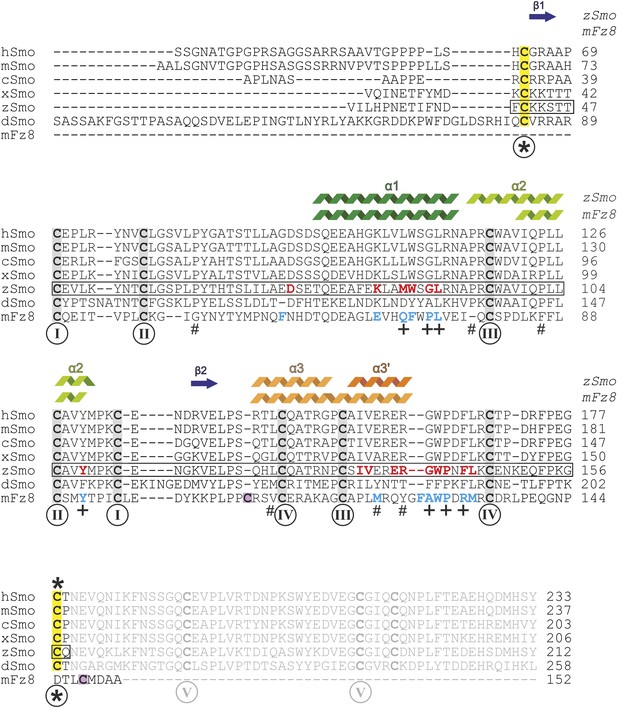
Sequence alignment of the ectodomains of Smo family members and the CRD of mFz8.
Sequences were aligned using ClustalW (Larkin et al., 2007) and adjusted manually for mFz8. Secondary structure assignments of zSmo CRD and mFz8 (PDB ID 4F0A, Janda et al., 2012) are displayed above the alignment and color-coded as in Figure 5. Disulfide bonds are highlighted and numbered as in Figure 5A. Smo disulfide bond *, which is not conserved in the CRD protein family, is marked in yellow. The two cysteine residues of mFz8 forming the rearranged disulfide bond (marked with * in Figure 5C) are highlighted in violet. The box indicates the zSmo residues visible in our crystal structure. Residues lining the oxysterol binding groove in Smo are highlighted in red for zSmo and residues lining the palmitoleyl-binding groove in mFz8 are in blue. Mutated mSmo residues that substantially reduced binding to 20(S)-OHC beads are depicted with a plus (+) below the alignment. Mutated residues that did not reduce binding are marked with a number sign (#).
Structural comparison of CRDs.
https://doi.org/10.7554/eLife.01340.012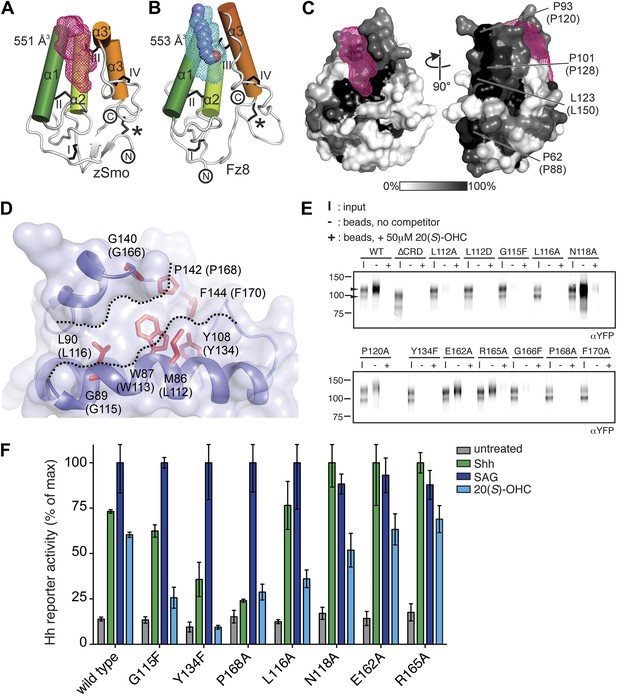
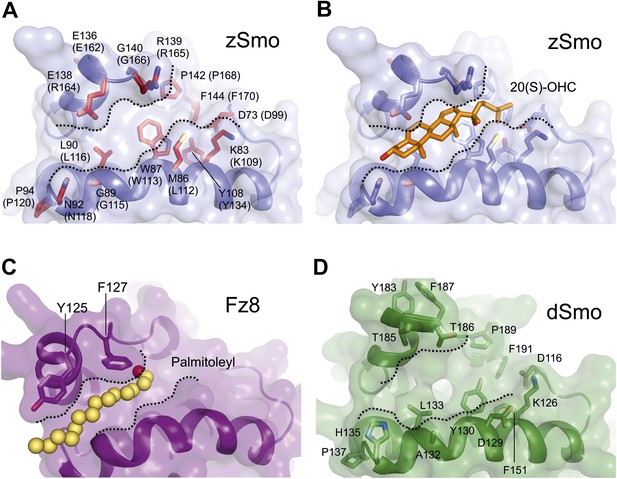
Mapping and analysis of the zSmo oxysterol binding site.
(A and B) Ribbon representations of the zSmo CRD (A) and the Fz8 CRD-palmitoleyl (B) structures. View and presentation follows Figure 5A. The palmitoleyl-binding pocket of Fz8 is depicted as a cyan wire mesh and the corresponding pocket in the zSmo CRD structure is in red wire mesh. Volumes were calculated using the program Volumes (RE Esnouf, unpublished), with a 1.4 Å probe radius. The palmitoleyl moiety is shown as slate spheres. (C) The solvent accessible surface of the zSmo CRD is color-coded according to residue conservation (from non-conserved, white, to conserved, black) based on alignments containing amino acid sequences from >80 vertebrate Smo proteins. The right panel is rotated 90° around the y-axis relative to the left panel. Residues on the opposite face of the oxysterol-binding pocket that were subjected to mutagenesis are labeled. (D) Close-up view of the potential 20(S)-OHC binding site in the zSmo CRD structure. Residues predicted to make contacts with 20(S)-OHC are shown in stick representation and highlighted in red. Boundaries of the hydrophobic groove are marked with dotted lines. zSmo residues are numbered, with the corresponding mSmo residues in parentheses. (E) The indicated full-length mSmo point mutants were tested for their interaction with 20(S)-OHC beads in the absence or presence of free 20(S)-OHC competitor. Well-folded Smo mutants ran as a double band on a 4–12% Bis-Tris gradient gel (arrowheads), with only the slower-migrating species being captured on 20(S)-OHC beads. (F) A Hh reporter assay was used to measure signaling in Smo−/− cells transiently transfected with constructs encoding various mSmo point mutants and then treated (48 hr) with Shh, SAG (100 nM) or 20(S)-OHC (10 μM). The maximum reporter response for each mutant was set to 100%. Error bars denote S.D. (n = 3).
Molecular modeling analysis of the zSmo CRD.
(A) Close-up view of the Smo oxysterol-binding groove. Presentation is as in Figure 6D. Boundaries of the potential binding site are marked with dashed lines. (B) The computationally docked structure of 20(S)-OHC (stick representation; carbon: orange, oxygen: red) in complex with the zSmo CRD suggests energetically favorable interactions between the two molecules with an estimated free binding energy of −9.0 kcal/mol and estimated inhibition constant, Ki, equal to 260 nM. View is as in (A). (C) Close-up view of the palmitoyl binding site in the Fz8-Wnt complex (PDB ID 4F0A, Janda et al., 2012) in the same orientation as in (A). (D) Homology model of the Drosophila Smo-CRD (dSmo) based on the zSmo structure (sequence identity: 42%) reveals a substantially wider groove compared to grooves of the CRDs from zSmo and Fz8. Three key residues (Met86, Tyr108, Gly140) are absent in dSmo that have been shown to be essential for 20(S)-OHC binding to vertebrate Smo.
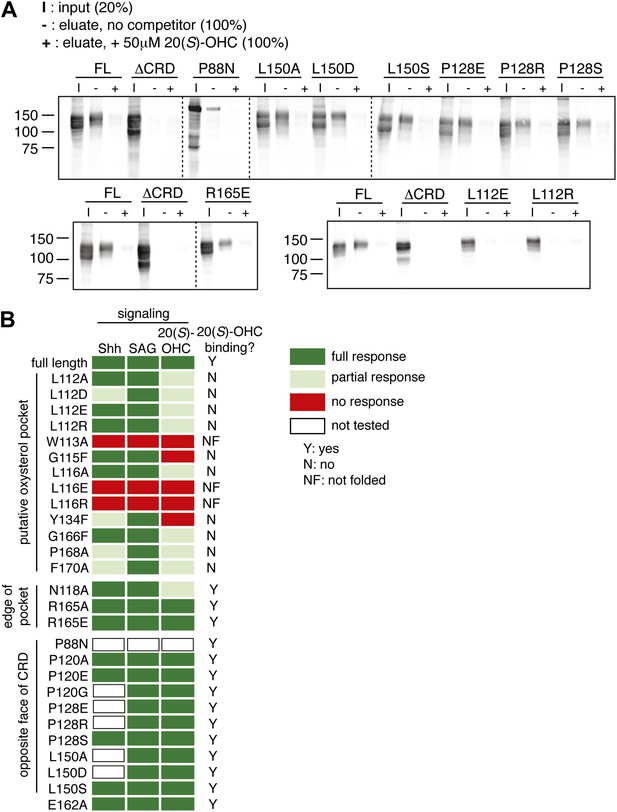
Mutagenesis of the putative oxysterol binding site in the mSmo CRD.
(A) The indicated full-length mSmo point mutants were tested for their interaction with 20(S)-OHC beads in the absence or presence of free 20(S)-OHC competitor. (B) Heat map summarizing the signaling properties of all Smo mutants tested in this study in response to Shh, SAG and 20(S)-OHC, using assays of the type shown in Figure 6E,F. Mutants that were not responsive to SAG and/or ran as a single band on an SDS-PAGE gel (indicating lack of glycan chains attached in the Golgi apparatus) were deemed not folded (NF). Mutants were only assessed for Shh and 20(S)-OHC responsiveness if they had SAG responsiveness that was at least 75% of that seen with wild-type mSmo.
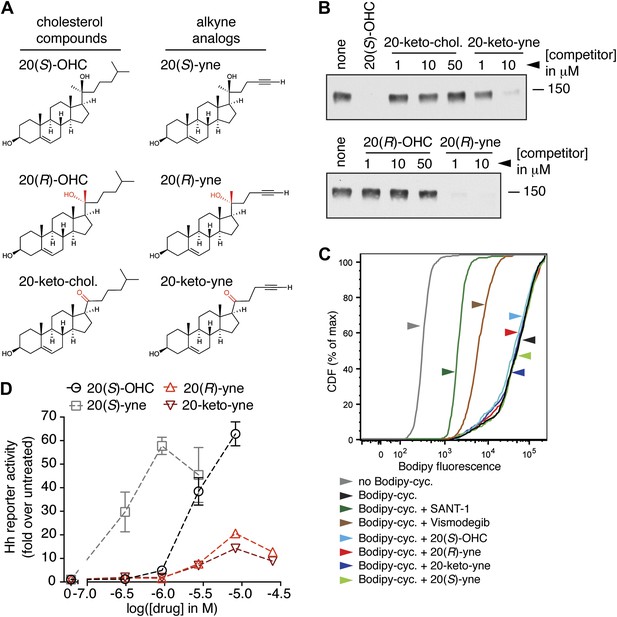
Partial agonists that target the Smo CRD.
(A) Structure and synthetic logic for 20(R)-yne and 20-keto-yne. 20(R)-OHC and 20-keto-cholesterol are related analogs that lack the alkyne moiety. (B) Immunoblots show the amount of mSmo CRD-Fc captured on 20(S)-OHC beads in the presence of the indicated oxysterols added as competitors. (C) Binding of bodipy-cyclopamine to cells expressing full-length mSmo was determined by FACS in the presence of various Smo ligands. The bodipy-cyclopamine fluorescence in a cell population is expressed as a cumulative distribution function (CDF), which denotes the percentage of cells that show a given level of fluorescence or lower. Bodipy-cyclopamine binding can be competed by SANT-1 and Vismodegib, two 7TM site ligands, but not by any of the CRD-binding oxysterols. (D) Hh reporter activity in cells treated with increasing concentrations of the indicated oxysterols.
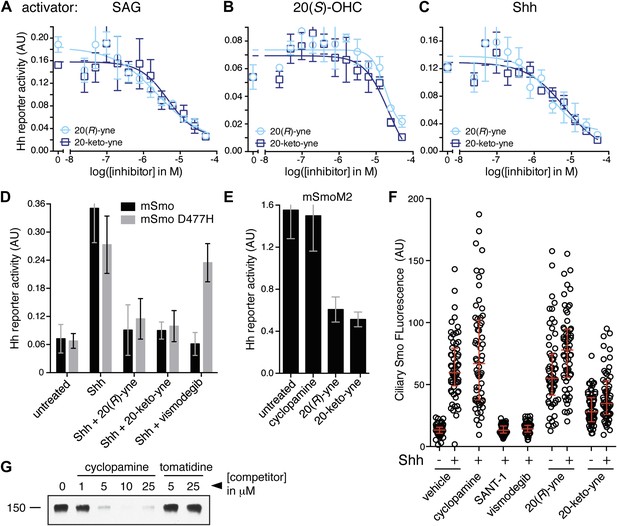
20(R)-yne and 20-keto-yne can inhibit Hh signaling.
(A–C) Hh reporter activation by 100 nM SAG (A), 5 μM 20(S)-OHC (B) and Shh (C) can be inhibited by both 20(R)-yne and 20-keto-yne. (D) Hh reporter activity in Smo−/− cells transfected with wild-type mSmo or mSmo D477H and then treated (48 hr) with Shh in the presence of 20(R)-yne, 20-keto-yne (both at 25 µM) or vismodegib (100 nM). (E) Hh reporter activity in NIH 3T3 cells transfected with constitutively active, oncogenic mSmoM2 and then treated (12 hr) with 20(R)-yne and 20-keto-yne (10 µM each) or cyclopamine (1 µM). (F) Accumulation of endogenous mSmo in cilia of NIH 3T3 cells treated (4 hr) with the indicated Smo ligands in the presence or absence of Shh. Each point represents the Smo fluorescence in a single cilium and the red lines denote the median and the interquartile range of mSmo fluorescence (n = 60 for each condition). (G) The binding of mSmo CRD-Fc to 20(S)-OHC beads can be inhibited by cyclopamine but not by the structurally-related alkaloid tomatidine. Error bars denote S.D. (n = 3).
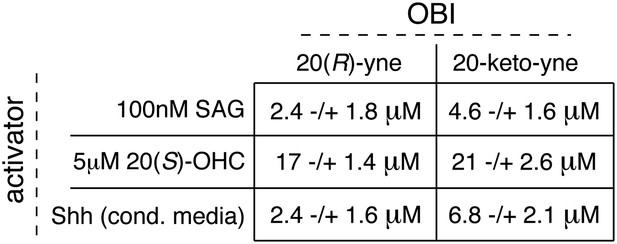
Table of IC50 values for the OBIs.
In Figure 8A,B,C, we demonstrate the inhibitory activity of 20(R)-yne and 20-keto-yne in the presence of three different Hh activators (SAG, 20(S)-OHC and Shh). Using curve fitting (described in detail in methods), we derived IC50 values for each inhibitor in the presence of the three different activators. Values represent mean ± SEM.
Tables
Data collection and refinement statistics
| SeMet-substituted zSmo CRD | Native zSmo CRD | |
|---|---|---|
| Data collection | ||
| Beamline | ESRF-ID23-EH1 | DIAMOND I03 |
| Wavelength | 0.979 | 1.000 |
| Space group | P43212 | P43212 |
| Cell Dimension (Å) | a, b = 68.2; c = 92.3 | a, b = 68.6; c = 95.3 |
| Resolution | 48.0–2.3 (2.36-2.30) | 31.0–2.6 (2.67-2.60) |
| Completeness (%) | 99.2 (92.8) | 98.9 (94.7) |
| Unique reflections | 10,085 | 7307 (491) |
| Rmerge (%) | 10.0 (92.0) | 13.4 (64.8) |
| I/σ(I) | 25.5 (3.9) | 14.4 (2.4) |
| Multiplicity | 26.5 (23.7) | 8.8 (6.0) |
| Refinement | ||
| Resolution range (Å) | 30.50–2.30 | 31.00–2.60 |
| No. reflections | 9576 | 7275 |
| Rwork (%) | 23.6 | 21.6 |
| Rfree (%) | 28.4 | 26.0 |
| No. atoms (protein/Zn/Na/water) | 1880/1/3/28 | 1880/1/2/48 |
| B-factors (Å2) (protein/Zn/Na/water) | 57/60/43/48 | 40/32/27/30 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.012 | 0.004 |
| Bond angles (°) | 1.604 | 0.714 |
| Ramachandran statistics | ||
| Favored (%) | 96.5 | 97.9 |
| Disallowed (%) | 0.4 | 0 |
-
Each structure was determined from one crystal. Numbers in parentheses refer to the highest resolution shell. Rfree equals the R-factor against 5% of the data.





