An allosteric Sec61 inhibitor traps nascent transmembrane helices at the lateral gate
Figures
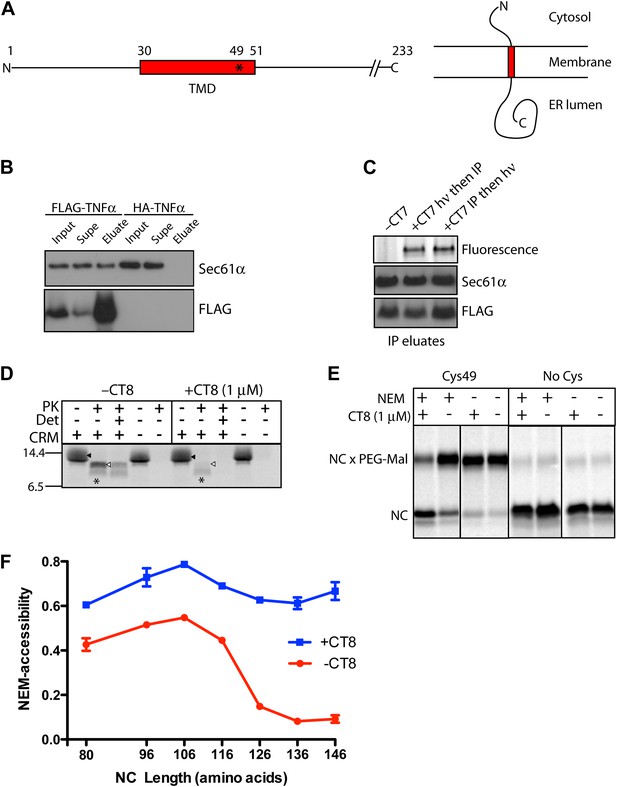
CT8 blocks TMD integration into the membrane.
(A) Schematic of TNFα primary structure and membrane orientation. Cys49 is indicated by an asterisk. (B) TNFα 126-mers containing an N-terminal FLAG- or HA-tag were translated in the presence of canine rough microsomes, solubilized with 1% Deoxy BigChap (DBC), and immunoprecipitated (IP) with anti-FLAG affinity resin. IP eluates were analyzed by immunoblotting for FLAG-TNFα and Sec61α. (C) As in (B), except that microsome-targeted 126-mers were prepared in the presence or absence of the photo-affinity probe CT7 and photolyzed (hν) either before (lane 2) or after immunoprecipitation (lane 3). The photo-crosslinked CT7/Sec61α adduct was detected by click chemistry with TAMRA-azide, followed by in-gel fluorescence imaging. (D) Protease protection assays with 35S-labeled 126-mers translated in the presence or absence of canine rough microsomes (CRM), followed by treatment with proteinase K (PK) and TX-100 (Det) as indicated. The nascent chain and protease-protected fragment are indicated (closed and open triangle, respectively). An unidentified weak band (asterisk) was occasionally observed. (E) Microsome-targeted 126-mers with Cys49 or lacking all cysteines were assembled in the presence or absence of CT8 and treated with N-ethylmaleimide (NEM) as indicated. Samples were subsequently denatured with SDS, treated with PEG-maleimide (PEG-Mal), and analyzed by SDS-PAGE/phosphorimaging. NEM inaccessibility is indicated by a shift to higher molecular weight, corresponding to nascent chains (NC) modified by PEG-Mal at Cys49. (F) NEM accessibility of RNCs of varying length, defined as fraction of nascent chains modified by PEG-Mal, determined by phosphorimaging and normalized to a matched control reaction lacking NEM. Data represent the mean ± range of two independent experiments. For raw phosphorimaging data, see Figure 1—figure supplement 1C.
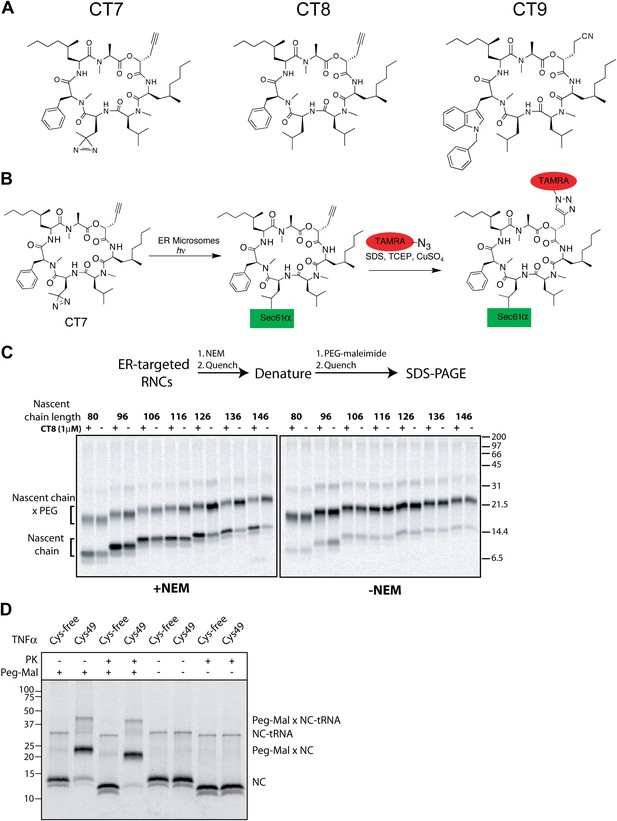
CT8 blocks TMD integration into the membrane.
(A) Chemical structures of cotransin variants used in the study. CT7 contains a photo-reactive diazirine to enable covalent photo-crosslinking to Sec61α. All compounds block TNFα insertion into ER microsomes with similar potency (Maifeld et al., 2011, and data not shown). (B) Scheme for CT7 photo-affinity labeling of Sec61α. The covalent CT7/Sec61α adduct (formed after irradiation with UV-light, hν) is detected using click chemistry between the alkyne group in CT7 and a rhodamine-azide (TAMRA-N3). (C) NEM accessibility assays of Cys49 RNCs of varying length in the presence and absence of CT8. Gels are representative of the primary data used to derive the graph in Figure 1F. (D) Microsome targeted TNFα 126-mers with Cys49 or lacking all cysteines were treated with proteinase K (PK) as indicated. Samples were subsequently denatured with SDS, treated with PEG-maleimide (PEG-Mal) as indicated and analyzed by SDS-PAGE/phosphorimaging. PEG-Mal efficiently modifies the PK-protected TNFα fragment containing Cys49.
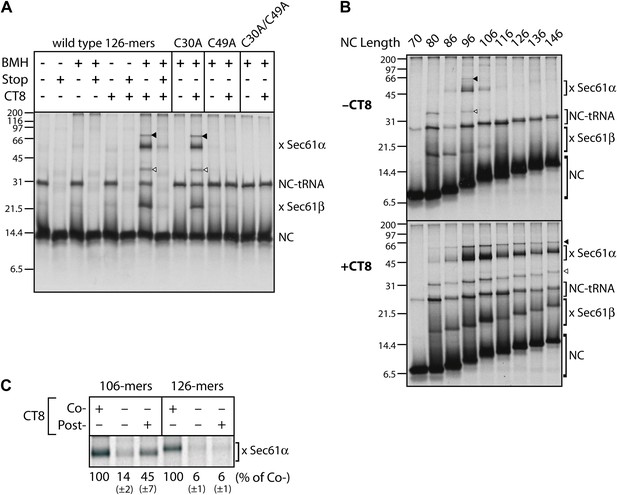
CT8 stabilizes a transient pre-integrated intermediate.
(A) Microsome-targeted TNFα 126-mers assembled in the presence or absence of CT8 (1 μM) were treated with BMH as indicated (Stop: termination codon at position 126). Bands corresponding to the nascent chain (NC) and the NC crosslinked to Sec61α and Sec61β are indicated (for IP confirmation, see Figure 2—figure supplement 1). Residual NC-tRNA as well as crosslinks to Sec61α and Sec61β is also indicated (closed and open triangle, respectively). (B) BMH crosslinking reactions with TNFα RNCs of varying length. (C) Microsome-targeted TNFα RNCs of the indicated length were assembled in the continuous presence of CT8 (‘Co-’) or treated with CT8 post-translationally (‘Post-’), followed by BMH crosslinking. NC crosslinks to Sec61α were quantified by phosphorimaging and normalized to the cotranslationally CT8-treated control. Quantified data represent the mean ± standard deviation of three independent experiments. In (B) and (C), TNFα RNCs contained a single cysteine (Cys49).
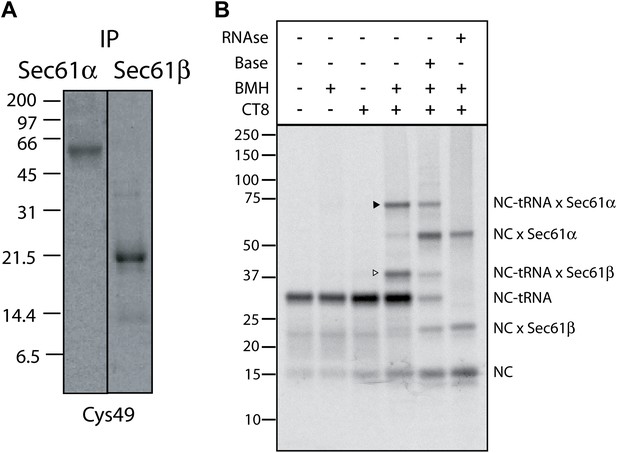
CT8 stabilizes a transient pre-integrated intermediate.
(A) Denaturing immunoprecipitation (IP) of BMH crosslinking reactions. Cys49 126-mers were prepared in the presence or absence of CT8 and crosslinked with BMH. Reactions were then quenched with DTT, denatured in SDS and immunoprecipitated with antibodies directed against the indicated proteins. Eluates were analyzed by SDS-PAGE and autoradiography. (B) Microsome targeted wild-type TNFα 126-mers were assembled in the presence or absence of 1 µM CT8 and treated with BMH as indicated. The BMH treated samples were further treated with base or RNAse A and reactions analyzed on NuPage gels at pH 7.3. Base and RNAse treatment collapses the nascent chain-tRNA bands.
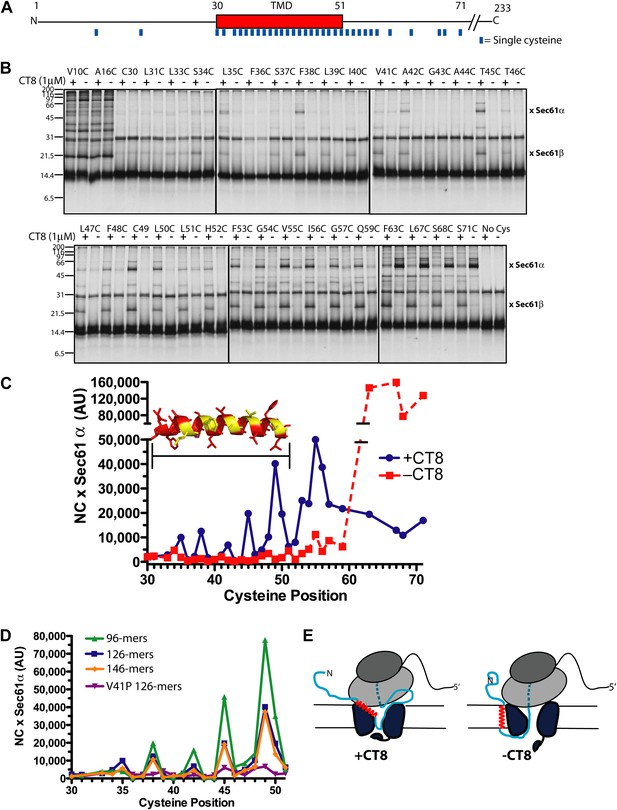
CT8 traps the TMD in a helical conformation with a defined orientation.
(A) Positions of single cysteine mutations (blue dashes) analyzed by BMH crosslinking. (B) BMH crosslinking reactions of 126-mers containing a single cysteine at the indicated positions. (C) Crosslinking intensities (NC × Sec61α) were quantified by phosphorimaging of the gels shown in (B) and plotted as a function of cysteine position. Inset, TMD α-helix model (red) highlighting positions with strongest crosslinks to Sec61α (yellow). (D) Overlay of BMH crosslinking profiles for the indicated constructs. Crosslinking intensities were normalized to an internal standard (Cys49 126-mer) included in each experiment. For raw phosphorimaging data, see Figure 3—figure supplement 1. (E) Cartoon depicting approximate TMD disposition (red) of 126-mer assembled in the presence and absence of CT8.
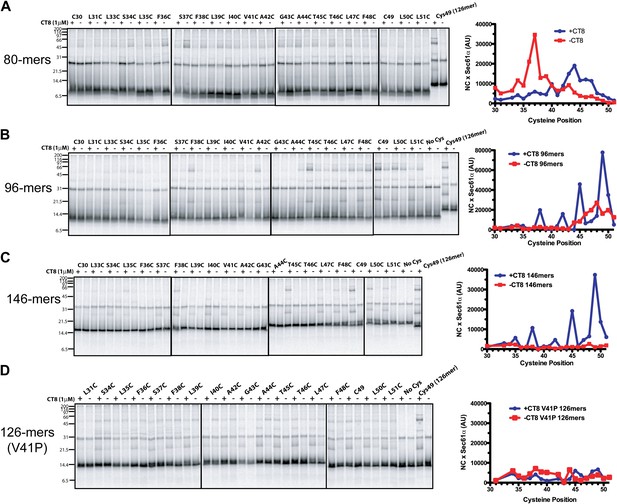
CT8 traps the TMD in a helical conformation with a defined orientation.
(A–D) BMH crosslinking profiles of RNCs of various lengths that carry single cysteines at the indicated positions of the TMD. The graphs on the right plot the intensity of the TMD/Sec61α crosslink (×Sec61α) against the position of the cysteine along the TMD. For part (D), the V41P position was not changed to cysteine. An internal standard construct (Cys49 126-mer) was included in each experiment (last pair of lanes in each gel series) to enable comparison of crosslinking efficiencies between different experiments. The values on the y-axes for plots in (A–D) are therefore comparable.
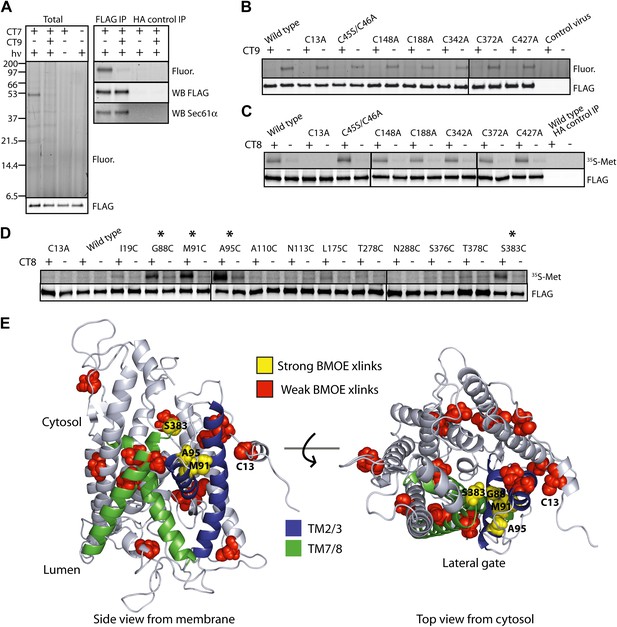
The pre-integrated TMD docks to the cytosolic tip of the lateral gate.
(A) Cotransin binding activity of recombinant FLAG-Sec61α. Microsomes from Sf21 cells expressing FLAG-Sec61α and Sec61γ were incubated with CT7 (50 nM) in the presence or absence of excess CT9 (10 μM). Samples were photolyzed (hν), and analyzed as in Figure 1C (left panel). Samples were immunoprecipitated under denaturing conditions with anti-FLAG or anti-HA (control) beads and analyzed as previously (right panel). (B) Photo-affinity labeling with CT7 as in (A) with microsomes from FLAG-Sec61α/γ mutant or control baculovirus-infected Sf21 cells. (C) 35S-labeled TNFα 126-mers (Cys49) were translated in the presence of FLAG-Sec61α/γ microsomes and subjected to BMH crosslinking. After immunoprecipitation with anti-FLAG beads, eluates were analyzed by autoradiography and immunoblotting. (D) As in (C), except that crosslinking reactions were performed with BMOE instead of BMH. See Figure 4—figure supplement 1 for further characterization of recombinant FLAG-Sec61α/γ. (E) Homology model of human Sec61α (Erdmann et al., 2009) (based on PDB entry:1RH5), highlighting positions of native and engineered cysteines (yellow = strong, red = weak crosslinks to TNFα TMD Cys49). Lateral gate helices are shown in blue (TM2b/3) and green (TM7/8).
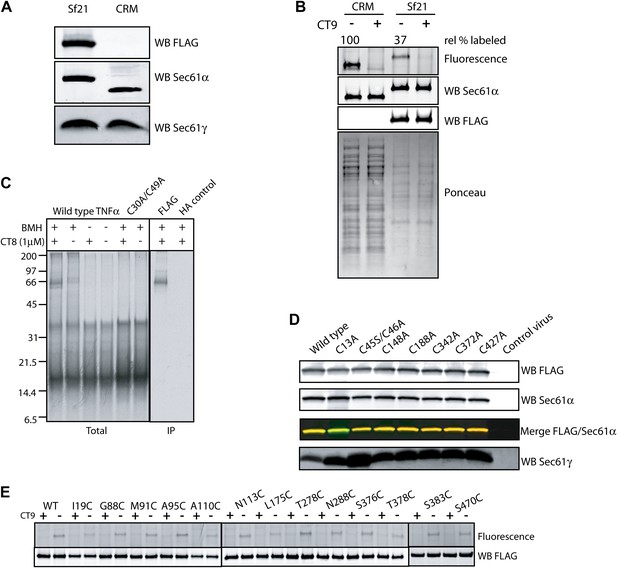
The pre-integrated TMD docks to the cytosolic tip of the lateral gate.
(A) Co-immunoprecipitation of recombinant FLAG-Sec61α and Sec61γ. Sf21 insect cell microsomes were solubilized with 1% Deoxy BigChap (DBC) and subjected to anti-FLAG immunoprecipitation (IP). Eluted samples (Sf21) were analyzed by immunoblotting next to a sample of canine microsomal ER membranes (CRM). (B) Comparison of the relative CT7 photo-crosslinking efficiency of canine rough microsomes (CRM) and Sf21 microsomes containing wild-type FLAG-Sec61α/γ complex. CT7 crosslinking was performed as before. The relative crosslinking efficiency of recombinant Sec61α was normalized to the native protein. (C) BMH crosslinking reactions using recombinant FLAG-Sec61α/γ complex. Cys49 TNFα 126-mers were prepared in the presence of Sf21 microsomes, and BMH crosslinking reactions were conducted as described previously. Reactions were either analyzed directly (Total) or after denaturing IP with anti-FLAG affinity resin. IP with the anti-HA affinity resin served as a specificity control. (D) Western blots of Sf21 microsomes co-expressing either wild-type or mutant FLAG-Sec61α/γ. Insect cell microsomes from cells infected with a baculovirus that lacked Sec61 genes (control virus) served as a control. (E) CT7 crosslinking of mutant Sec61α/γ complexes containing the indicated cysteine substitutions. The region of the gel corresponding to FLAG-Sec61α is shown. Detection of photo-affinity labeling that is competed by CT9 indicates that the mutant Sec61α/γ complex is properly folded.
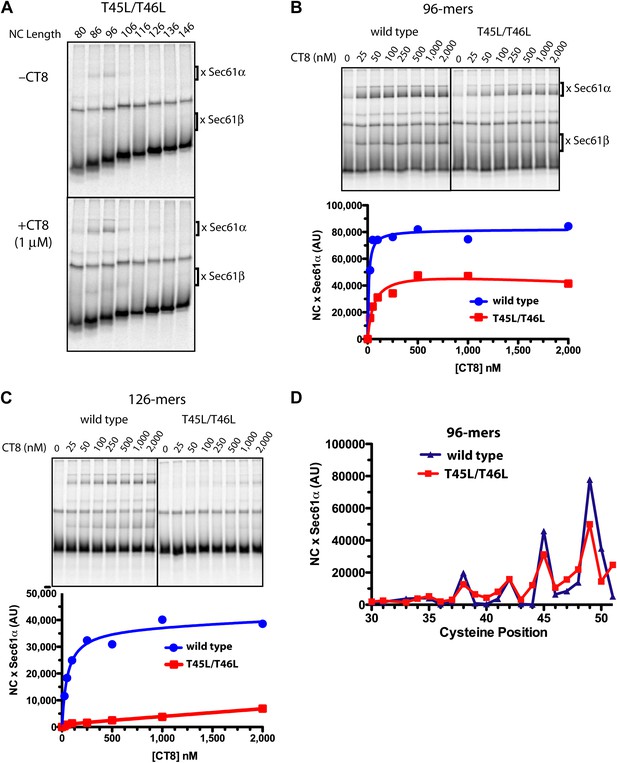
Resistant and sensitive TMDs pass through a common pre-integrated intermediate.
(A) BMH crosslinking reactions with T45L/T46L TNFα RNCs of varying length, assembled in the presence or absence of CT8. (B) BMH crosslinking reactions of wild-type or T45L/T46L 96-mers in the presence of increasing concentrations of CT8. Crosslink intensities (× Sec61α) were quantified by phosphorimaging and plotted as a function of [CT8]. (C) As in (B), except 126-mers were used. (D) T45L/T46L 96-mers containing single cysteines at the indicated positions were assembled in the presence of CT8 (1 μM) and subjected to BMH crosslinking. Crosslink intensities (× Sec61α) were quantified by phosphorimaging (see Figure 5—figure supplement 1 for raw data) and plotted as a function of cysteine position. Data from ‘wild-type’ 96-mer cysteine scan (Figure 3D) are shown for comparison.
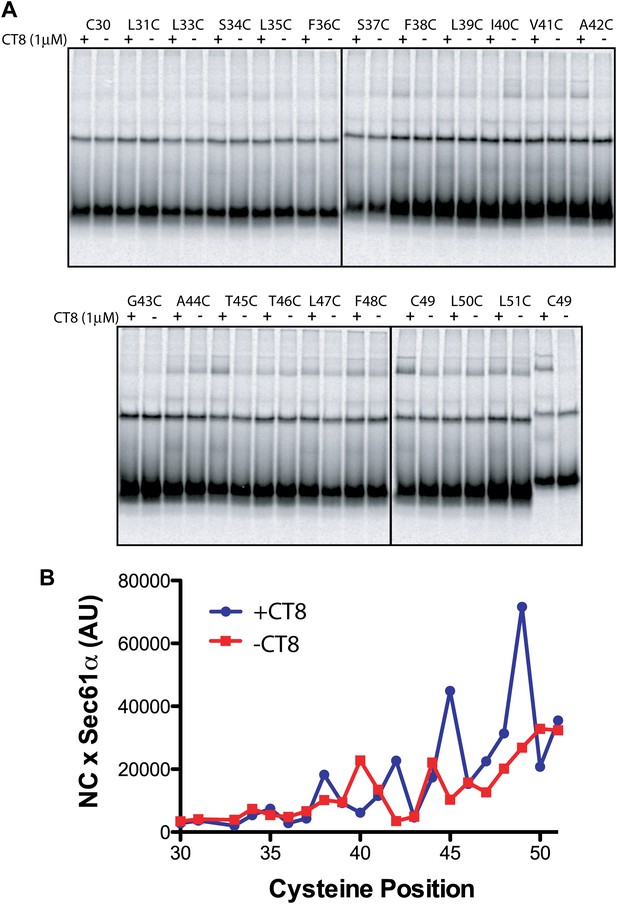
Resistant and sensitive TMDs pass through a common pre-integrated intermediate.
(A) BMH crosslinking reactions of T45L/T46L 96-mers containing single cysteines at the indicated positions. (B) Quantified data from the gels shown in part (A).
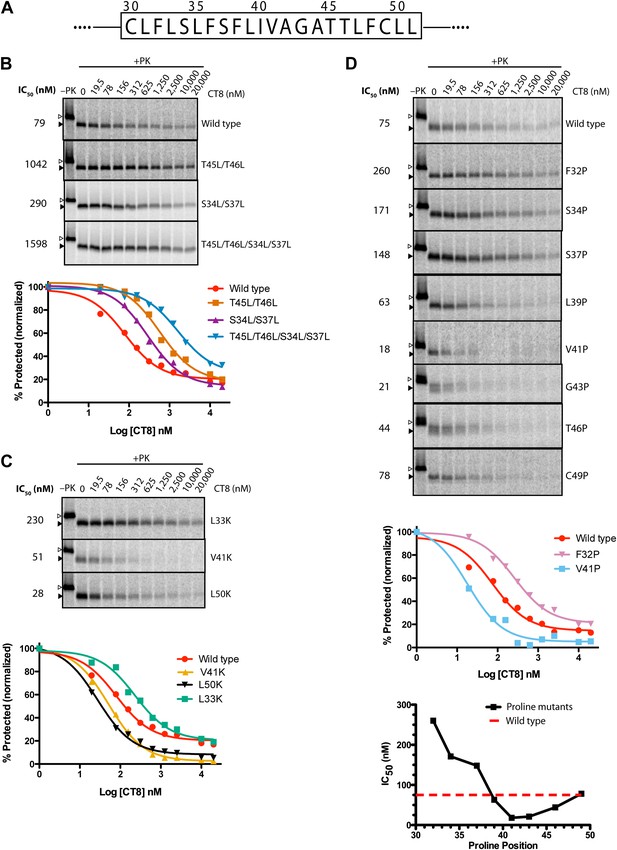
TMD sequence and biophysical properties determine CT8 sensitivity.
(A) Amino acid sequence of the TNFα TMD. (B) Protease protection assays (proteinase K, PK) of full-length TNFα polar-to-leucine mutants as a function of increasing CT8 concentration. Translation reactions were carried out in the presence of increasing concentrations of CT8 or DMSO control, followed by digestion with PK as previously described (Sharma et al., 2010). Full-length TNFα and the protease-resistant fragment are indicated by open and closed triangles, respectively. (C) As in (B), except with hydrophobic-to-lysine mutants. (D) As in (B), except with proline-scanning mutagenesis. IC50 values were plotted as a function of the position of the corresponding proline mutation. To compute IC50 values, protease-resistant fragments were quantified by phosphorimaging, normalized to the DMSO control, and fitted to a three-parameter equation using GraphPad Prism software. Representative curve fits in each set (B–D) are shown. Based on the online TMD prediction algorithm (http://dgpred.cbr.su.se/), no mutation was expected to change the N-terminal or C-terminal boundaries of the TMD.
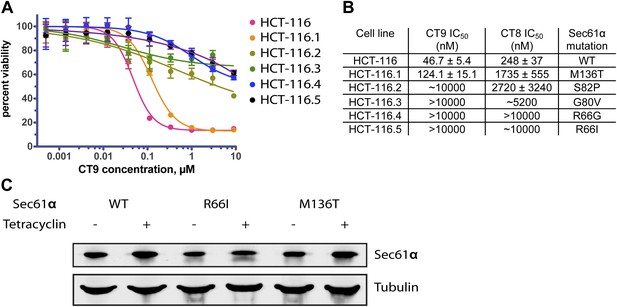
Mutations in the plug/lateral gate interface confer resistance to cotransin.
(A) HCT-116 parental cells and resistant clones were treated with increasing concentrations of CT9 for 72 hr, and viability was assessed by the Alamar Blue assay (mean ± S.D., n = 4). (B) Sensitivity of wild-type and resistant HCT-116 cells to CT8 and CT9 and the identified heterozygous S61α mutations. (C) Untagged Sec61α constructs are expressed in a tetracycline-dependent manner similarly as endogenous Sec61α.
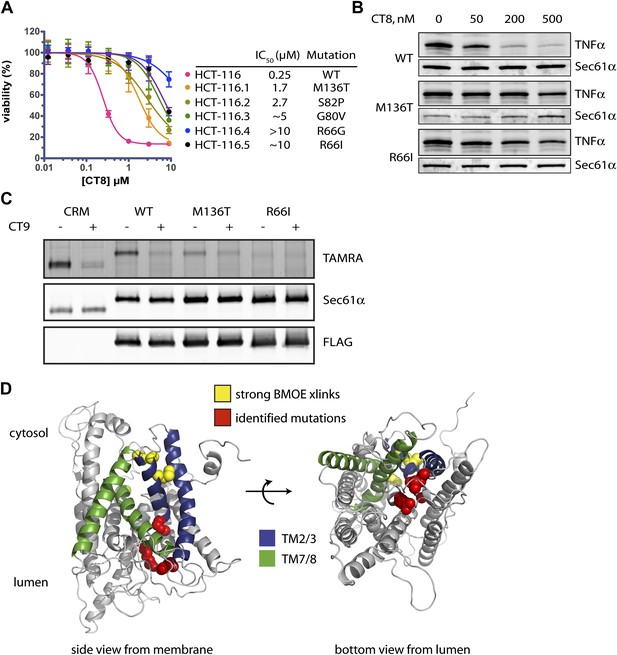
Mutations in the lumenal plug of Sec61α confer resistance to cotransins.
(A) Parental HCT-116 cells and resistant clones were treated with increasing concentrations of CT8 for 72 hr, and viability was assessed by the Alamar Blue assay (mean ± S.D., n = 4). (B) HEK293-FRT cells stably expressing wild-type or mutant Sec61α were transfected with a plasmid encoding TNFα and treated with increasing concentrations of CT8. After 24 hr, TNFα expression was analyzed by immunoblotting. (C) CT7 photo-crosslinking to recombinant wild-type and mutant FLAG-Sec61α. Microsomes were incubated with 250 nM CT7 in the presence or absence of excess CT9 (10 µM), photolyzed, and analyzed by in-gel fluorescence and immunoblotting as in Figure 1C. (D) Homology model of human Sec61α showing the location of cotransin resistance mutations (red) and the TMD docking site (yellow). Lateral gate helices colored as in Figure 4E.
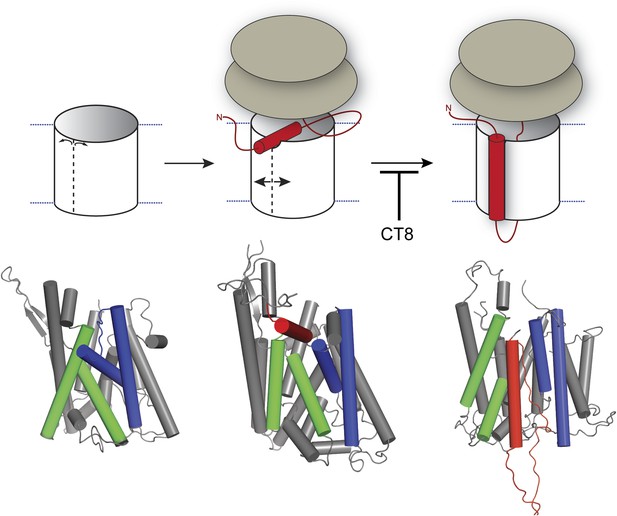
Model for cotransin-mediated inhibition of TMD integration.
Following RNC targeting to Sec61 (left panel), the TMD docks to a notch in the cytosolic tip of the lateral gate created by the separation of TM2b from TM8 (middle panel). This otherwise transient, pre-integrated configuration is stabilized by CT8 binding to the lumenal plug region of Sec61α. Interactions between the TMD and the lateral gate facilitate gate opening and TMD movement to a second site on the lipid-exposed face of the lateral gate (right panel). CT8 blocks TMD movement from the cytosolic docking site to the external, lipid-exposed site. Structural models (bottom) were adapted from PDB entries 1RH5 (left panel, closed lateral gate), 2ZJS (middle panel, pre-integrated configuration), and 3J00/3J01 (right panel, integrated configuration). The model for the pre-integrated TMD configuration was approximated by manually placing the TMD (modeled as an α-helix) into the cytosolic vestibule between TM2b and TM7 (PDB entry: 2ZJS), consistent with the BMOE crosslinking data. Lateral gate helices TM2b/3 and TM7/8 are blue and green, respectively, while the TMD helix is red.





