Crystal structures of virus-like photosystem I complexes from the mesophilic cyanobacterium Synechocystis PCC 6803
Abstract
Oxygenic photosynthesis supports virtually all life forms on earth. Light energy is converted by two photosystems—photosystem I (PSI) and photosystem II (PSII). Globally, nearly 50% of photosynthesis takes place in the Ocean, where single cell cyanobacteria and algae reside together with their viruses. An operon encoding PSI was identified in cyanobacterial marine viruses. We generated a PSI that mimics the salient features of the viral complex, named PSIPsaJF. PSIPsaJF is promiscuous for its electron donors and can accept electrons from respiratory cytochromes. We solved the structure of PSIPsaJF and a monomeric PSI, with subunit composition similar to the viral PSI, providing for the first time a detailed description of the reaction center and antenna system from mesophilic cyanobacteria, including red chlorophylls and cofactors of the electron transport chain. Our finding extends the understanding of PSI structure, function and evolution and suggests a unique function for the viral PSI.
https://doi.org/10.7554/eLife.01496.001eLife digest
Photosynthesis—the process by which plants and other organisms harness the energy in sunlight—is the source of almost all oxygen, food and fuel on earth. Oxygenic photosynthesis in living cells involves a series of reactions catalyzed by large protein complexes, various other soluble chemicals, and the transfer of electrons from so-called donors to acceptors. The energy in the sunlight is captured by two membrane-embedded protein complexes—photosystem I, which is the most powerful electron donor in nature, and photosystem II—and converted into chemical energy.
Almost half of the world’s photosynthesis occurs in the oceans, and is performed by single-celled cyanobacteria and algae. Interestingly, some of the genes that encode photosynthetic enzymes in cyanobacteria are also found in the genomes of viruses that infect these bacteria. It is thought that these viruses can alter photosynthetic pathways in their hosts, but the interactions between these viruses and their hosts are not fully understood.
Now, Mazor et al. have created a photosystem I complex that mimics the viral version of this complex, and have gone on to solve its three-dimensional structure. This mimetic virus-encoded complex was shown to be a ‘promiscuous’ electron acceptor: this means that, unlike most electron acceptors, it can accept electrons from more than one electron donor.
Further, Mazor et al. solved the structure of photosystem I from Synechocystis, a cyanobacterium that lives in fresh water; and found some surprising differences between it and the only other published structure for photosystem I from a cyanobacterium (which was from a species that lives in hot water springs). These included differences in components involved in the electron transfer chain—a series of chemical reactions in which electrons are passed from donor to acceptor molecules—that were thought to be highly conserved. Other differences in the structures allowed Mazor et al. to identify the location of a unique chlorophyll pigment group in the Synechocystis photosystem I.
Since Synechocystis is commonly used as a model to study photosynthesis, an improved understanding of its photosystem I should lead to further improvements in our knowledge of photosynthesis.
https://doi.org/10.7554/eLife.01496.002Introduction
Oxygenic photosynthesis, which takes place in cyanobacteria, algae, and plants, provides most of the food and fuel on Earth (Barber, 2004; Nelson, 2011). Cyanobacterial photosynthetic membranes contain two photosystems, of which PSII mediates the extraction of electrons from water, the initial electron donor, to the plastoquinone pool. PSI mediates electron transfer from cytochrome C553 (or plastocyanin in eukaryotes) to ferredoxin, thereby generating the reducing power needed for CO2 fixation in the form of NADPH. The architecture of oxygenic photosynthesis in cyanobacteria has largely been determined and the structures of both photosystems from thermophilic cyanobacteria have been solved at high resolution (Jordan et al., 2001; Ferreira et al., 2004; Nelson and Ben-Shem, 2004; Amunts et al., 2007; Umena et al., 2011).
Photosynthetic reaction centers are classified according to their terminal electron acceptor as either type I, with an iron sulfur cluster acceptor, or type II, with a quinone terminal acceptor. PSI, a type I reaction center, presumably evolved from the simpler homodimeric bacterial reaction centers (Buttner et al., 1992; Liebl et al., 1993; Baymann et al., 2001; Nelson and Ben-Shem, 2005). We currently know only two versions of the type I reaction centers: the relatively simple bacterial homodimer found in green sulfur bacteria (GSB), heliobacteria and Chloracidobacterium or the much more complex PSI with its 11–15 subunits found in cyanobacteria and all photosynthetic eukaryotes (Buttner et al., 1992; Hauska et al., 2001; Nelson and Yocum, 2006). Although sequence conservation between the PsaA/B subunits of PSI and the single large subunit of the homodimeric reaction centers is low, their inferred membrane topology is similar, and the sequence conservation around the bound iron sulfur cluster is high, supporting the notion of a common ancestor (Mulkidjanian and Junge, 1997; Blankenship and Hartman, 1998; Baymann et al., 2001; Mazor et al., 2012; Rutherford et al., 2012).
In the earth ocean’s, mainly single cell organisms, cyanobacteria or algae, perform oxygenic photosynthesis. One of the most intriguing aspects of marine cyanobacterial photosynthesis is the involvement of various viruses in the process. Hints on this involvement come from the presence of photosynthetic genes in viral genomes (Mann et al., 2003; Sullivan et al., 2006). Many marine phages encode for either the D1 gene or both D1 and D2 which together make up the reaction center of PSII (Sullivan et al., 2006). In addition, several subunits of the NDH1 complex and soluble cytochromes are also found in the viral metagenome (Philosof et al., 2011). The presence of these genes suggests that marine viruses have the ability to alter the host photosynthesis pathways in ways, which are still not completely understood. Recently, an operon encoding a complete, albeit minimal, PSI complex was found in the genome of several marine cyanophages (Sharon et al., 2009). The viral PSI has a unique gene composition compared to the PSI composition of its hosts. PsaL and PsaI are missing and a unique PsaJ-PsaF (PsaJF) fusion-protein exists.
The absence of both PsaL and PsaI from the viral PSI suggests that it functions as a monomer and does not assemble trimers since both PsaL and PsaI are important for the formation of the cyanobacterial trimer (Chitnis and Chitnis, 1993; Xu et al., 1995). In eukaryotes, the PsaF subunit is important for electron transfer between plastocyanin (PC) and the PSI complex (Farah et al., 1995; Hippler et al., 1996). Although the structures of PsaF from cyanobacteria and plants are very similar, the role of PsaF is less well defined in cyanobacteria (Chitnis et al., 1991; Xu et al., 1994a, 1994b; Hippler et al., 1996). Structurally, the N-terminus of PsaF forms a major ‘bump’ in the flat, luminal side of PSI. Within this domain, the eukaryotic PsaF contains several additional positively charged residues that stabilize the interaction between PSI and PC (Farah et al., 1995; Hippler et al., 1996). In the viral PSI the N-terminus of PsaF is deleted and as a result the luminal face of PSI is expected to be relatively flat.
The unique light harvesting and transfer requirements, together with the large number of interconnected subunits, have resulted in a very high degree of structural conservation in PSI complexes (Ben-Shem et al., 2003). For example, approximately 90 chlorophyll molecules, bound by the PSI reaction center remained largely unmoved by more than 2 billion years of evolution between the thermophilic cyanobacterium Thermosynechococcus elongatus and the flowering plant Pisum sativum (Jordan et al., 2001; Ben-Shem et al., 2003; Amunts et al., 2007, 2010). A common feature of PSI is the presence of low energy chlorophyll molecules called ‘red chlorophylls’. Red chlorophylls are present in the reaction center of cyanobacteria and in the light-harvesting belt of the plant PSI. In cyanobacteria, red chlorophylls can affect the trapping kinetics of excitation energy as shown by measurements done on PSI from various species (Gobets et al., 2001). The location of the red chlorophylls within the internal antenna of PSI is still a matter of debate, the inter species variability of the red absorption suggests that they are located in a relatively variable region of PSI. Currently, all the calculations of individual pigment energies are based on the high resolution PSI structure from Thermosynechococcus elongatus, which contains a relatively large number of red pigments. New, high resolution structures are needed to compare antenna system across various species.
In this work, we characterized both biochemically and structurally a phage-mimetic PSIPsaJF constructed in Synechocystis. We find that the fused subunit PsaJF is incorporated into the PSI complex and that this incorporation results in a promiscuous PSI, able to accept electrons from respiratory cytochromes. We also present a high resolution model for the Synechocystis PSI monomer, which allows us to identify for the first time the changes in the antenna system and electron transport chain (ETC) that exist between Synechocystis and Thermosynechococcus. The location of red chlorophylls in the thermophilic vs the mesophilic cyanobacteria is discussed.
Results
Structure of a trimeric PSIPsaJF
We hypothesized that the consequences of this viral JF gene product would be a PSI complex with more promiscuous electron acceptor reactions (Sharon et al., 2009). Cyanobacteria employ a multitude of electron transfer reactions in their membrane systems. These systems are at least partially insulated by spatial confinement of the various electron donors and acceptors, but the molecular interactions between these donors and acceptors may also contribute to pathway isolation. To explore the promiscuity of viral PSI complexes, we constructed a phage-mimetic PsaJ-PsaF fusion in the easily cultivable cyanobacterium Synechocystis sp. PCC 6803. The borders of PsaJ and PsaF are easily located in the viral gene and the high conservation of the PsaF subunit enabled us to duplicate the salient features of the viral gene into our artificially constructed fusion made from the corresponding Synechocystis genes (Figure 1—figure supplement 1). Synechocystis strains carrying the fused PsaJF gene as their sole source of PsaF and PsaJ, grow at wild type rates in heterotrophic conditions and without added carbon. After purifying the PSI complex from the photosynthetic membranes of both wild-type and PsaJF strains, we found that the trimer to monomer ratio is similar in both strains, indicating that the assembly and stability of PSIPsaJF is not disturbed (Figure 1A). The subunit composition of the purified complex showed that it indeed contained the fused PsaJF subunit in lieu of PsaJ and PsaF (Figure 1B).
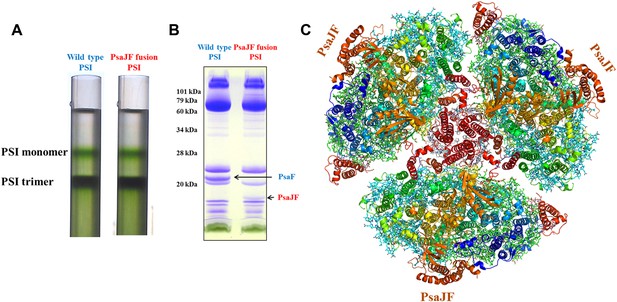
The crystal structure of a PsaJF fusion containing PSI complex.
(A) Sucrose gradients of chlorophyll containing complexes from either wild type or PsaJF containing bacteria showing similar trimer to monomer ratios in both strains. (B) An SDS-PAGE gel showing the polypeptide composition of the trimeric PSI fractions from the sucrose gradient showing the presence of the PsaJF fusion (which was confirmed by MS analysis) in the purified PSI complex. (C) The PSI trimer structure viewed from the stromal side of the membrane showing the fused subunit assembled in the complete complex.
To get a more detailed view of the PsaJF subunit in PSIPsaJF, we crystallized PSIPsaJF and obtained diffraction data to 3.8 Å resolution (Table 1). The crystal structure of the PSIPsaJF complex was solved by molecular replacement, first by using the previously determined cyanobacterial structure as a search model (PDB ID 1JBO) and later with a higher resolution model from Synechocystis (see ‘Results’ below). The model reveals three monomers in the asymmetric unit. A complete trimeric complex is formed from monomers positioned in three unit cells (Figure 1C). As seen in Figure 2A,B, the overall architecture of PSI from Synechocystis is highly similar to the Thermosynechococcus elongatus PSI with two noticeable exceptions, the fused PsaJF subunit and PsaX, which as expected, is missing (the genome of Synechocystis PCC6803 does not contain a PsaX homolog).
X-ray data collection and refinement statistics
| Data collection | PsaJF trimer | PSI monomer | |
|---|---|---|---|
| Beamline | ESRF–ID29 | ESRF–ID29 | SLS–PXI–X06SA |
| Wavelength (Å) | 0.97625 | 0.97625 | 1 |
| Resolution (Å) | 30–3.8 | 30–2.8 | 30–3 |
| Measured reflections | 426,209 (63,882) | 419,672 (61,558) | 396,647 (57,225) |
| Unique reflections | 113,221 (16,536) | 91,895 (13,209) | 72,095 (10,585) |
| Rpim (%) | 6.8 (71) | 7.5 (124) | 5.6 (77.3) |
| I/σ(I) | 9.9 (1.2) | 8 (1.3) | 10 (1.2) |
| Completeness | 98.9 (99.5) | 99.4 (99.1) | 96.8 (98.3) |
| Redundancy | 3.8 (3.9) | 4.6 (4.7) | 5.1 (5.4) |
| Space group | P 21 | P 21 21 21 | P 21 21 21 |
| Unit cell dimensions | |||
| a, b, c (Å) | 214, 134, 220 | 120, 173, 179 | 120, 174, 179 |
| α, β, γ (°) | 90, 111.1, 90 | 90, 90, 90 | 90, 90, 90 |
| Refinement statistics | |||
| Resolution (Å) | 30–3.8 | 30–2.8 | 30–3 |
| Rwork/Rfree | 25.9/29.7 | 21/24 | 24.4/28 |
| No. of chains | 30 | 9 | 9 |
| No. of ligands | 360 | 119 | 116 |
| Average B-factor (Å2) | 128 | 85 | 90 |
| R.M.S deviations | |||
| Bond Angles | 1.7 | 1.7 | 1.7 |
| Bond lengths | 0.014 | 0.005 | 0.004 |
| Ramachandran statistics | |||
| Favored region % | 93.8 | 93.8 | 93.2 |
| Allowed region % | 5.1 | 5.9 | 6.3 |
| Outlier region % | 1.1 | 0.3 | 0.5 |
| clashscore | 5.5 | 3.8 | 4.6 |
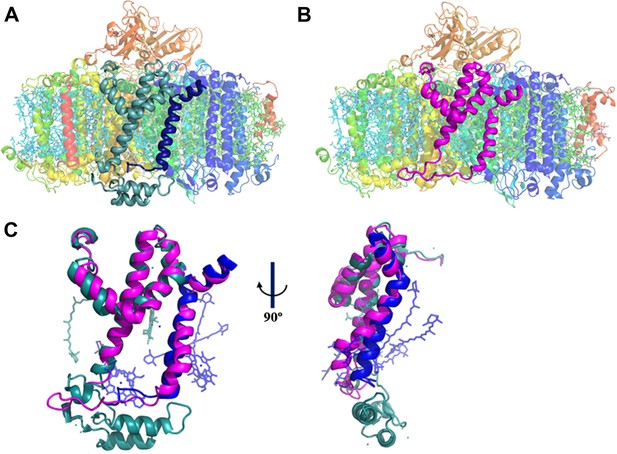
A JF containing PSI complex has a luminal plateau.
(A) PSI from Thermosynechococcus with PsaJ (Blue) and PsaF (cyan) shown from the membrane plane, the N-terminus of PsaF extends as two alpha helices into the luminal space. PsaX is colored red. (B) The PsaJF (magenta) containing PSI complex from Synechocystis showing the flatter luminal plateau created from the removal of the N-terminus of PsaF. (C) Superposition of the JF subunit from Synechocystis (magenta) on the PsaF (cyan) and PsaJ (blue) subunits from Thermosynechococcus, the overall structure of the fused subunit remains undisturbed.
Our trimeric PSI structure is the first look at a mesophilic cyanobacterial PSI. As such, even at this relatively low resolution, some interesting features are apparent. The structure clearly shows the presence of the fused subunit integrated in the PSIPsaJF complex (Figure 2B). Deleting the N-terminus of PsaF resulted in a flattened plateau on the luminal side of the complex (Figure 2B,C). Three chlorophyll molecules and four carotenoids are coordinated by PsaJ and PsaF in the Thermosynechococcus PSI structure (Figure 2C). We find that all of these ligands are probably lost during the assembly of PSIPsaJF and could not be traced in our model (Figure 2C). In spite of the relatively low resolution, we find that the overall organization of the other cofactors in PSI is hardly affected in PSIPsaJF demonstrating the mild effect of the JF mutation on the overall organization and integrity of the complex.
PSIPsaJF is a promiscuous electron acceptor
Next we tested whether PSIPsaJF is a promiscuous electron acceptor. We measured the half-life of PSI reduction by a mammalian respiratory cytochrome c. As seen in Figure 3A, P700 in PSIPsaJF is reduced much faster by a respiratory cytochrome c than in the wild-type complex. However, PSIPsaJF is reduced by its native electron donor cytochrome c553 (CytC553) with similar kinetics compared to the wild-type PSI (Figure 3B). This is in contrast to eukaryotic PSI where the N-terminus of PsaF was shown to be important for the electron donation reaction between plastocyanin and PSI (Hippler et al., 1996). The similar electron donation rates observed with CytC553 are in agreement with previously published results obtained from PsaF and PsaJ deleted strains (Xu et al., 1994a, 1994b). When the light-dependent oxidation of cytochrome c was followed we obtained similar results. Compared to the wild-type complex, PSIPsaJF had faster cytochrome oxidation kinetics when respiratory cytochrome was used (Figure 3C), and similar kinetics when native CytC553 was used (Figure 3D). The similarity between the reaction kinetics of PSI reduction together with the similar cellular growth rates of wild-type and PSIPsaJF strains shows that deleting the N-terminus of PsaF causes no gross perturbation to the interaction between PSI and CytC553, that is, the N-terminus of PsaF does not play any positive role in this interaction in Synechocystis. The N-terminus of PsaF can act as a negative regulator of some electron donation reactions, especially from positively charged donors such as respiratory cytochromes. We suggest that this was the original function of PsaF in a more primitive form of PSI and only later, as PSI evolved, the binding site was refined to its final configuration. In the viral PSI, such promiscuity can be beneficial when the viral reaction center encounters the highly variable cytochromes found in marine cyanobacteria (Mazor et al., 2012) or alternatively to allow PSI to accept electrons from respiratory activities when PSII activity is compromised.
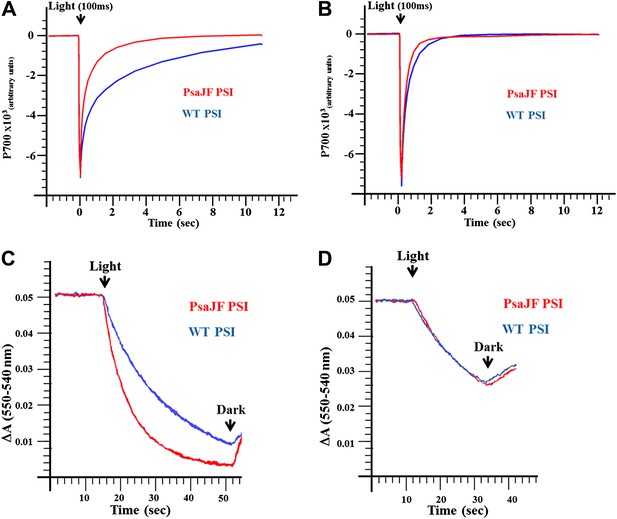
PSIPsaJF is a promiscuous electron acceptor.
(A) P700 reduction kinetics (after a 100 ms pulse of orange light) showing faster reduction kinetics of a PSIPsaJF complex with respiratory CytC (red line) compared to the wild-type complex (blue line). (B) Native cytochrome C553 reduces both wild-type and PSIPsaJF complexes at similar rates. (C) Plots of the cytochrome oxidation (ΔA 550 nm–540 nm) under continuous white illumination by PSI, showing faster oxidation of a respiratory cytochrome by PSIPsaJF (red trace) compared to the wild-type PSI (blue trace). (D) Native CytC553 is oxidized with similar kinetics by both complexes.
High-resolution structure of a PSI monomer
The small subunits PsaF, J, K, L, I, and M of the PSI complex can each be deleted without observing any severe growth defects in cells (Chitnis and Chitnis, 1993; Xu et al., 1995; Naithani et al., 2000). Their conservation in all cyanobacterial species, arguably the most diverse group of microorganisms, suggests that over evolutionary time periods losing them is detrimental. Different small subunits, however, are functionally connected. For example, in cells lacking PsaJ, PsaF is only partially incorporated into the complex (Xu et al., 1994a). A similar situation is seen in PsaI deleted cells, where PsaL is only partially assembled into the complex (Xu et al., 1995). In both the cases, a ‘simple’ single transmembrane (TM) helix (PsaJ or PsaI) seems to be necessary for the assembly of a more complex subunit (PsaF or PsaL, respectively).
To improve the diffraction from our crystals, and increase the observation to parameter ratio of our model, we purified and crystallized the monomeric form of PSI. This approach was attempted previously, but did not bear fruit (Jekow et al., 1995, 1996; Fromme and Mathis, 2004). We inserted 10 histidine residues at the C-terminus of PsaL, similar to an approach previously shown to completely disrupt trimer formation without adversely affecting PSI activity (Tang and Chitnis, 2000). As expected, extending the C-terminus of PsaL caused no significant effect on cell growth or PSI activity and resulted in a homogenous population of monomeric complexes that were readily purified and crystallized (Figure 4—figure supplement 1A,B).
Fortuitously, PsaL and PsaI were missing from the final purified complex (Figure 4—figure supplement 1C), making the subunit composition of the crystallized PSI very similar to the viral PSI complex (the only difference is the single TM subunit PsaM which is found in our preparation and is absent from the viral PSI operon). This shows that the subunit composition of the phage complex is sufficient to produce a stable and active PSI complex.
The crystals obtained from this monomeric form of PSI belong to a higher symmetry space group (P212121), had a reduced cell volume (∼3.75 × 106 Å3) and, most importantly, diffracted to a resolution of 2.8 Å. At this resolution, the entire complement of ligands and cofactors is revealed, and it is feasible to carry out a detailed comparison between the thermophilic and mesophilic complexes.
Electron transport chain
The available structures of PSI from thermophilic cyanobacteria and higher plants show complete conservation in all the components of the electron transport chain (ETC) from P700 to the iron sulfur clusters (Jordan et al., 2001; Ben-Shem et al., 2003; Amunts et al., 2007). In light of this complete conservation, we were surprised to discover a large rearrangement of the isoprenoid tail of the phylloquinone (Q1A) coordinated by PsaA (Figure 4B, Figure 4—figure supplement 2). This movement is accompanied by rearrangement of the phytol tails of chlorophylls A06 and A40 (ligands are named according to Jordan, et al. whenever possible [Jordan et al., 2001]). In spite of these large movements, the positions of the quinone heads and chlorine rings are highly conserved between all known PSI structures (Figures 4B and 5A). The movements in Q1A and the surrounding chlorophylls appear to originate from two amino acid changes that occurred in the PsaJ subunit between Synechocystis and Thermosynechococcus. Ala J16 in Thermosynechococcus changed to Met J16 in Synechocystis, making contacts with terminal carbon atoms of Q1A (Figure 4C). Similar contacts are seen in the Thermosynechococcus Q1A terminal carbon atoms, but these contacts are made with Met 19 in PsaJ, which changed to Leu 19 in Synechocystis (Figure 4C). Interestingly, this interaction between the terminal carbon atoms of the isoprenoid tail and methionine is present in a number of cyanobacterial species (Figure 4—figure supplement 4). It is interesting to note that most of the other contacts stabilizing the tail conformation of Q1A mainly involve interactions between the hydrophobic tails of other ligands.
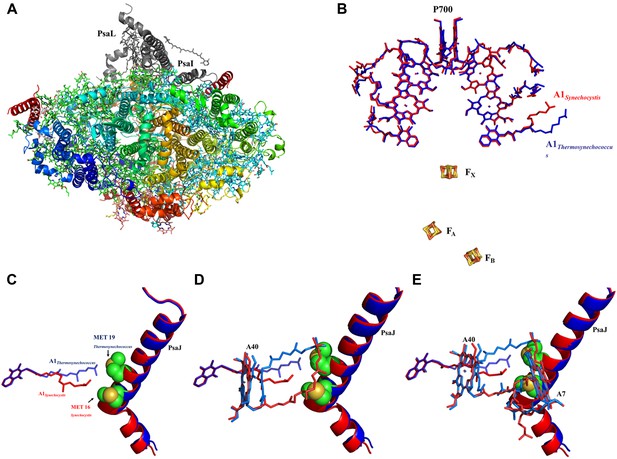
High resolution structure of a monomeric PSI complex from Synechocystis.
(A) A luminal view of the monomeric PSI complex. PsaL and PsaI that are missing from our crystal are shown in gray. (B) Comparison of the ETC of Synechocystis (red) to that of Thermosynechococcus (blue) reveals near perfect superposition in all the components with the clear exception of the quinone A1 isoprenoid tail. (C) Superposed PsaJ from Synechocystis (red) and Thermosynechococcus (blue) showing the coordinating methionine residue (carbons in green and the sulfur atom in yellow). (D) The phytol tail of chlorophyll A40 is reoriented in Synechocystis in order to accommodate the movement of the quinone tail. (E) The phytol tail of Chlorophyll A1 in Synechocystis shifted to accommodate the PsaJ MET16 change.
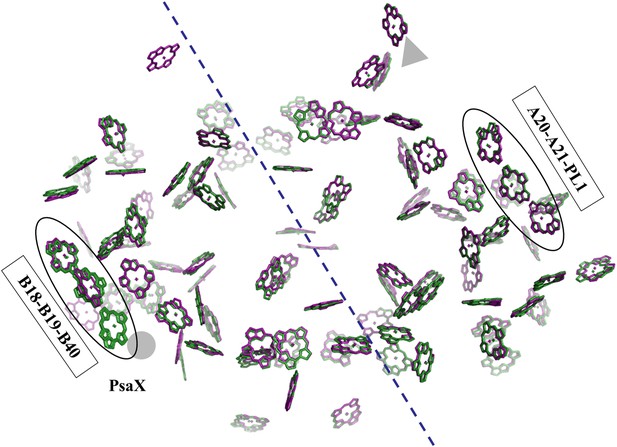
Pseudo C2 symmetry in the antenna system of Synechocystis.
A depth cued image comparing the overall organization of the PSI antenna from Synechocystis (green) or Thermosynechococcus (purple) viewed from the stromal side of the membrane, only chlorine rings are shown for clarity. The C2 pseudosymmetry axis is shown in blue. PsaX is shown as gray filled circle. The new chlorophyll detected in Synechocystis (B40) is clearly seen next to PsaX which precludes its binding in Thermosynechococcus.
Another noticeable feature of the ETC is the conservation of water molecules position around Fx (Figure 4—figure supplement 3). This water pocket is positioned between Q1B and Fx and can potentially modify the ET rates between the A and B branches of PSI. Mutations in the coordinating tryptophan residue greatly affect the ET rates between Q1B and Fx (Ali et al., 2006).
Changes in the orientation of the isoprenoid tail can fine-tune the ETC. The fact that small, less conserved, subunits, such as PsaJ, are involved in changing these orientations, provides relaxed evolutionary constraints that facilitate faster exploration of the available changes (Peisajovich et al., 2006). To summarize, the components of the ETC are extremely conserved over all known PSI structures. However, even small changes in the orientation of components can play a part in fine tuning ET processes. Most of the studies on the ETC are performed in Synechocystis (Savitsky et al., 2013) and assume that all of the factors involved occupy identical positions as shown in Thermosynechococcus or higher plants, this may lead to somewhat erroneous conclusions. The surprising observation of a conserved water cluster positioned along the ET path between Q1B and Fx suggests that this feature has a functional role.
Light-harvesting and red chlorophylls
The polypeptide chains of PSI coordinate more than a 100 pigments, which funnel excitation energy to P700 with near absolute quantum efficiency (Croce and van Amerongen, 2013). A common theme in PSI is the occurrence of red chlorophylls, which absorb (and fluoresce) light at longer wavelengths than P700 (Palsson et al., 1998; Gobets et al., 2001). Red chlorophylls slow the trapping kinetics of P700, however, the quantum yield of the complex is not affected by this at room temperature; at higher light intensities red traps may have a photoprotective role. In addition, red chlorophylls increase the absorbance cross section of the entire complex, especially in shaded environments.
There is considerable variability in the strength of the red absorption between various cyanobacterial species and in particular Synechocystis is known to have very low levels of red chlorophylls in comparison to Thermosynechococcus (Gobets et al., 2001). The exact identity of the red pigments within the reaction center of PSI is unknown and the clear difference in their content between Synechocystis and Thermosynechococcus presents us with an opportunity to identify these chlorophylls. Based on ring-to-ring distances and dipole orientation Jordan et al. 2001 identified one chlorophyll trimer (B31-B32-B33) and three dimers (A32-B7, A38-A37, B37-B38) as candidates for strongly coupled pigments in the reaction center. We find that the ring location and the local environment of dimer A38-A39 remained virtually unchanged between Synechocystis and Thermosynechococcus (Figure 5—figure supplement 1A,B). The chlorine rings of dimer B37-B38 occupy very similar positions in both photosystems including the coordinating water molecule of B37 that can clearly be seen in our maps (Figure 5—figure supplement 1C). The phytol tails of both chlorophylls shifted considerably, however because of this dimer’s proximity to PsaL definite conclusions are not possible (Figure 5—figure supplement 1C,D).
The ring positions of the A32-B7 dimer shifted slightly from Thermosynechococcus to Synechocystis, and the side chain coordinating the magnesium of B7 changed from glutamine to histidine. These changes can contribute greatly to the Qy position of the pigment (Wientjes et al., 2012). However, this dimer is also located close to PsaI and PsaL, and therefore may have been affected by their exclusion from the complex (Figure 5—figure supplement 1E,F). Indeed, in the PSIPsaJF model, the positions of both rings are slightly shifted toward the position observed in Thermosynechococcus. This dimer may be sensitive to the oligomerization state of the complex, as it has been shown that some red absorption is lost upon monomerization (Gobets et al., 2001), a phenomenon which is common to both Synechocystis and Thermosynechococcus. A monomeric structure that includes both PsaI and PsaL will resolve this question.
In contrast, it is very clear that chlorophyll B33 is completely missing from the stacked trimer observed in Thermosynechococcus (Figures 5 and 6). This is accompanied by a small deletion in the PsaB loop, which supports B33. A similar deletion is found in approximately third of the cyanobacterial PsaB sequences, as well as the PsaB sequences in eukaryotes (Figure 6—figure supplement 1), where the corresponding chlorophyll has shifted substantially (Ben-Shem et al., 2003; Amunts et al., 2007). The ring distances and orientation between B31 and B32 hardly changed from Thermosynechococcus to Synechocystis. These findings strongly support the B31-B32-B33 trimer as the strong red absorber in Thermosynechococcus and suggest that the one of remaining dimers, either B37-B38 or B31-B32, is responsible for the residual red absorbance seen in Synechocystis.
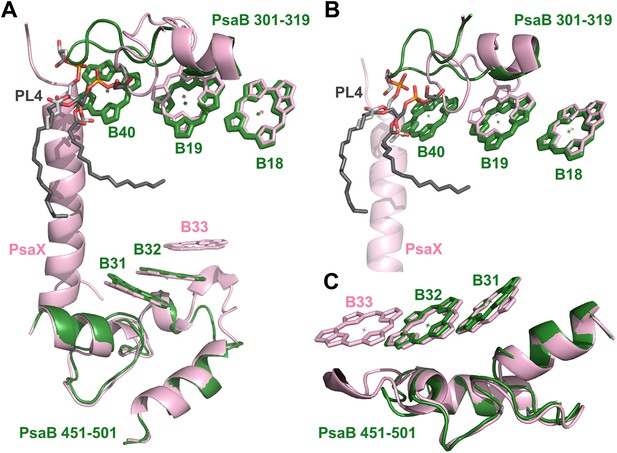
Complementing structural rearrangements on opposite sides of PSI in Synechocystis.
(A) The luminal-side chlorophyll trimer (B31-B32-B33) is missing in Synechocystis, while chlorophyll B40 forms a stromal chlorophyll trimer (Synechocystis in green, Thermosynechococcus in light pink). The PsaB loop on the stromal side (PsaB 301-319) extends to coordinate chlorophyll B40 and A phospholipid (shown in dark gray), while the luminal side PsaB loop (PsaB 451-501) is shortened in Synechocystis and chlorophyll B33 is lost as a result. (B) A close up on the stromal side changes showing the large conformational change in PsaB. In spite of this the phospholipid is clearly observed. (C) Loss of the B31-B33-B33 chlorophyll trimer in Synechocystis, the shortened PsaB loop together with the remaining chlorophylls in Synechocystis is shown.
Photosystem I contain another chlorophyll trimer that is coordinated by PsaA (A20-A21-PL1). The ring-to-ring distances between the chlorophylls of this trimer do not suggest direct coupling, but, surprisingly, our model revealed the presence of an additional chlorophyll trimer, related by the pseudo C2 symmetry axis of the reaction center (Figures 5 and 6A,B). The new trimer is formed by an additional chlorophyll, B40, which is coordinated by the head group of a phospholipid (one of three present in PSI) (Figure 6A,B). The phosphate oxygen atom lays 2.8 Å from the magnesium atom of B40. This coordination of chlorophylls by lipids is reminiscent of the situation in light-harvesting complex II (LHC-II) (Standfuss et al., 2005). In contrast, CP29, which is highly homologous to LHC-II, does not contain such a configuration and this probably contributes to the different non photochemical quenching properties of both proteins (Pan et al., 2011).
The position of this new chlorophyll (B40) is occupied by subunit PsaX in Thermosynechococcus (Figure 6). Surprisingly, and in spite of the large structural changes observed in this region, this lipid is still clearly visible, this high degree of conservation demonstrates the important role of the PSI lipids. The fact that the newly discovered chlorophyll trimer in Synechocystis relates to its counterpart by pseudo C2 symmetry (Figure 6) suggests that this configuration preceded the association of PsaX, which may have been an adaptation to high temperature. Possibly the most important role of PsaX is to protect this lipid at higher temperatures. The occurrence of this chlorophyll trimer suggests that these trimers have a structural role and the appearance of the red absorption results from fine-tuning of their position. It is likely that the pigment environment contributes significantly to its spectral properties.
Discussion
Photosystem I is one of the most complex enzymes in nature. Even though the evolution of photosynthesis must have taken a complex path, today we can only see a limited snapshot of this path. The high degree of sequence conservation and the importance of pigments to the photosynthetic process further limit our ability to deduce function from mere sequences.
Viral reaction centers
The prevalence of photosynthetic genes in marine cyanophage genomes suggests that photosynthesis and respiration play an important part in the phage–host interaction. Aside from the PSI operon, genes for other subunits of the major complexes of the photosynthetic electron transport chain reside in phages including PSII, NDH-I, F-ATPase (Philosof et al., 2011). Small soluble proteins like cytochromes and ferredoxins are also abundant (Sharon et al., 2011). A common property of these genes is that they cluster together as a distinct group when compared to their cyanobacterial counterparts (Zeidner et al., 2005). In some cases, it is possible to suggest that viral subunits modify the flow of electrons in the photosynthetic chain based on the known function of the cyanobacterial counterpart. This is the case with the virally encoded NDH-I subunits, where the function of the native, bacterial complex in cyclic electron flow is established.
In this study, we show that a phage specific mutation, removal of the N-terminus of PsaF, results in a promiscuous PSI complex. It is known that the properties of the small electron donor (either PC or CytC553) largely determine its mode of interaction with PSI (Hervas et al., 2005). Marine cyanobacteria contain an especially diverse set of soluble cytochromes with variable physical properties (Mazor et al., 2012). How this diversity is manifested with respect to cellular respiration and photosynthesis is a matter for future experiments. However, our findings suggest that photosynthesis performed during phage infection and catalyzed (at least partially) by phage proteins (‘Viral photosynthesis’) utilize very different cytochromes efficiently and will not be sensitive to the nature of the electron donor. The existence of both NDH-I subunits and a promiscuous PSI operon in a single phage genome suggests that cyclic electron flow is enhanced during phage replication (Philosof et al., 2011). This can be achieved by increasing the flow of electrons into the photosynthetic electron transport chain through the NDH–I complex or by utilizing electrons from respiratory cytochromes or both. It is important to consider the two most common phage photosynthetic proteins, D1 and D2, in that light. The suggested function of virally encoded D1 and D2 is to replenish the rapidly damaged host D1 and D2 proteins during infection (Mann et al., 2003; Alperovitch-Lavy et al., 2011). This seems to partially contradict the notion of increased cyclic electron flow in infected cells but it is probable that some basal level of linear electron flow in the cells plays a major part in phage multiplication. Detailed studies performed in phage-infected cells are needed to uncover the facts needed to support these hypotheses.
We suggest that the phage-encoded PSI complex with its unique gene composition adds an important step to the scenario of PSI evolution from a primordial homodimeric entity to the current highly evolved complexes (Nelson, 2011; Mazor et al., 2012). In this scenario, the formation of the primordial heterodimer allowed for the evolution of the unique properties of the peripheral subunits, trimer formation in the case of PsaL and interaction with electron donors in the case of PsaF. The specificity and complexity of biological machines increases along their evolutionary path and PSI is a prime example of this property. Cyanobacterial PSI is much more complicated and intricate than its ancestral homodimer and the plant PSI adds additional levels of complexity over the cyanobacterial complex. The preservation of simpler forms of PSI is therefore quite unique and stems from the relatively simple life cycle of phages and from the unique evolutionary requirements associated with this life cycle. For example, some marine phages from the Myoviridae family can infect different species of cyanobacteria (Sullivan et al., 2003) and this creates unique requirements on their genes such as utilizing the different chlorophyll species found in Prochlorococcus and Synechococcus. The viral complex preserves some of the properties of the common PSI ancestor, lost from both the cyanobacterial and plant PSI. One of these properties appears to be promiscuity for electron donors as exemplified by the fused PsaJF subunit. The incredible richness uncovered by ocean genomic surveys suggests that further sampling will reveal further examples of ancestral forms of PSI and other unique versions of highly conserved genes.
Mesophilic cyanobacterial PSI
The two structures described in this work provide the first detailed look at the Synechocystis PSI complex. The two most prominent differences between Synechocystis and Thermosynechococcus are the longer trapping time for excitation energy and the stronger red absorbance found in Thermosynechococcus. Both of these differences are probably related to the number of red-shifted pigments found in PSI. This study provides definite proof for the absence of the putative ‘red’ chlorophyll trimer in Synechocystis and narrowed the number of coupled pigment dimers to two. Further calculations should help identifying other, more subtle alteration in pigment energy along the antenna system. Structurally, the Synechocystis PSI complex revealed the PsaX-less conformation of the antenna and it is evident that this configuration is closer to the plant PSI inner antenna and thus can be considered the ancestral arrangement.
One of the most surprising findings is the conserved lipid binding by the PsaB loop close to PsaX (Figure 6). This is the region of PSI that underwent the most drastic changes between Synechocystis and Thermosynechococcus, however the presence of the lipid is not affected by them. This suggests that the function of this lipid (and by inference the function of the other PSI lipids) is related to assembly process of PSI, possibly to support reinsertion into the membrane after the synthesis of an extended stromal loop. The absence of any sequence identifiable binding site for this lipid stems from the hydrophobic interactions made between its hydrocarbon tails and several chlorophyll molecules without involving any protein atoms. This highlights the significance of detailed structural information in elucidating the biology of complex biological assemblies.
A cluster of water molecules located between Fx and Q1B, conserved between Thermosynechococcus and Synechocystis was revealed. The two branches of the PSI ET chain are not equivalent, the PsaB located branch being faster. This water cluster may modulate the contribution by Tryptophan PsaB 664 to the electron transfer between Q1B and Fx. The high structural conservation observed in this region implies that the final answers can only be reached using ever more detailed models and refined calculation methods which will include all participating atoms.
Materials and methods
Culture conditions
Request a detailed protocolCyanobacteria were cultured in BG11 medium supplemented with 10 µg/ml Ferric ammonium citrate, 5 mM glucose and 10 µg/ml chloramphenicol under continuous white light (∼40 µE) in 30°C.
Strain construction
Request a detailed protocolSynechocystis PsaJ (amino acids 1–40) and PsaFΔN (amino acids 84–165) were amplified by PCR and fused together with a Cys-Ser-Cys linker between them. The PsaJF gene was fused to the native PsaF promoter together with 300 bp of upstream and downstream sequences to direct its recombination to the genome of Synechocystis. The Kanamycin resistance gene was cloned just downstream to the PsaJF gene. Primer sequences are available in Supplementary file 1.
Transformation of Synechocystis sp. PCC6803 was performed according to standard protocols. Gene replacement constructs were generated in pGEM or pJET, and used directly for transformation. All transformants were streaked on increased antibiotic concentrations for a minimum of three times, and verified by PCR.
Photosystem I purification
Request a detailed protocol20–40 l of cells were grown in regular BG11 (supplemented with 10 µg/ml ferric ammonium citrate and 5 mM glucose, plus 10 µg/ml chloramphenicol for strains with His-tagged PsaL) under light intensity of ∼40 µE at 30°C. Cells were harvested using centrifugation, and washed once by STN1 buffer (30 mM Tricine-NaOH pH 8, 15 mM NaCl, 0.4 M sucrose). Finally, cells were resuspended in 50 ml of STN1, and broken by an Avestin EmulsiFlex-C3 (three cycles at 1500 psi). The lysate was cleared by centrifugation in a SS34 rotor for 10 min at 12,000 rpm. Membranes in the supernatant were pelleted using ultracentrifugation (Ti70 rotor, 45,000 rpm for 2 hr), and resuspended in 50 ml STN2 (30 mM Tricine-NaOH pH-8, 100 mM NaCl, 0.4 M sucrose). After resuspension in STN2, the membranes were incubated on ice for 30 min, then collected again (Ti70 rotor, 45,000 rpm, 2 hr), and resuspended in approximately 15 ml of STN1. n-Dodecyl β-D-maltoside (DDM, Affymetrix, http://www.affymetrix.com/estore/index.jsp) was added to the membranes (from a 10% stock solution in water) at a 15:1 DDM to chlorophyll ratio. The suspension was gently mixed by hand a few times then incubated on ice for 30 min. After solubilization, the insoluble material was discarded using ultracentrifugation (Ti70, 45,000 rpm, 30 min). The solubilized membranes were loaded onto a DEAE column (DE52; Whatman http://www.gelifesciences.com/webapp/wcs/stores/servlet/catalog/en/GELifeSciences/brands/whatman/), column volume was adjusted to the chlorophyll content of the sample (1.5 or 1.2 ml DEAE per 1 mg chlorophyll for trimer or monomer preparations, respectively). The column was eluted using a linear NaCl gradient (15–200 mM NaCl for monomer preparation; 15–350 mM NaCl for trimer preparations) in 30 mM Tricine-NaOH pH 8, 0.2% DDM. Dark green fractions were collected and precipitated using PEG3350 (Hampton research http://hamptonresearch.com/Default.aspx), final PEG concentration was 8.5% for trimer preparations and 12% for monomer preparations. The green precipitate was resuspended in 30 mM Tricine-NaOH, pH 8, 0.05% DM, (10 mM NaCl for monomer; 75 mM NaCl for trimer), and loaded onto a 10–30% sucrose density gradient, prepared in 30 mM Tricine-NaOH pH-8, 0.05% DDM (10 mM NaCl for monomer; 75 mM NaCl for trimer). Following centrifugation (SW40 rotor, 37,000 rpm, 12 hr) the appropriate green band was collected and used for kinetic measurements or crystallization.
Photosystem I crystallization
Request a detailed protocolThe chlorophyll peak from the sucrose gradient was collected, NaCl was added to a final concentration of 150 mM and the complex was precipitated using PEG3350 (9% for trimeric complexes; 12% for monomeric complexes). After centrifugation (13,000 rpm, 5 min in an Eppendorf tabletop), the green precipitate was resuspended in minimal volume of buffer (3 mM Tricine-NaOH pH 8, 0.02% DDM), and any undissolved material was removed by repeating the centrifugation step. The chlorophyll concentration in the soluble material was adjusted to the desired concentration (typically 3 mg/ml) using the above buffer, and dispensed in 4 µl drops into 24-well, sitting drop crystallization plates. Protein drops were mixed 1:1 with reservoir solution (90 mM NaCl, 90 mM MgCl2, 0.0005% n-Nonyl-β-D-Maltoside (NM), 100 mM glycine, 30 mM Tricine-NaOH, pH 8, 9–11% gradient of PEG3350 for trimer complex; 60 mM Tricine-NaOH, pH 8, 9% PEG3350, 0.0005% NM, 100 mM glycine, 100–120 mM NaCl gradient for monomer complex). Crystals were formed within 1–3 days, and grown to their final size by a week. The crystals of the PSI trimer were large and flat with no precipitate in the well. In contrast, the monomeric complex formed multiple crystal forms. Only a small fraction of the crystals, recognized through their overall shape, diffracted to high resolution. The other crystals diffracted to very low resolution or did not diffract at all. Cryo protection was achieved by replacing the mother liquor in the well and in the reservoir with solutions containing progressively increasing PEG3350 concentrations (10%, 15%, 20% and 30%) with a minimum of 2 hr between each solution (usually 24 hr between each step). Finally crystals were placed in 40% PEG3350 solution for a few minutes, mounted on loops and frozen in liquid nitrogen.
Data collection and refinement
Request a detailed protocolData were collected at ESRF, Grenoble, station ID29, or at the SLS, Villigen, station PXI. Diffraction frames were processed with XDS (Kabsch, 2010), scaled and merged with SCALA (Evans, 2006; Winn et al., 2011).
Monomer
Request a detailed protocolTwo data sets from two different crystals were used in the refinement of the monomeric complex, one obtained from ID29 at the ESRF and one from PXI at SLS. Refinement was done with PHENIX (Adams et al., 2010). Monomer descriptions of chlorophyll and β-carotene were computed from average values of the 1.9 E PSII structure (Umena et al., 2011). At 3 Å the use of additional, external restraints was found to be ineffective. TLS refinement was used throughout the refinement. A very coarse choice of TLS groups was found to be optimal with the membrane domain, the stromal ridge and PsaK chosen as three separate TLS groups. Adding hydrogen atoms to the model was found to greatly improve the clash score and geometry of the model and so hydrogens were used throughout the refinement.
Trimer
Request a detailed protocolThe PSIPsaJF model was refined against a single data set obtained at ID29. Initially a truncated model (with most side chains and phytol tails removed) of PSI from Thermosynechococcus was used for molecular replacement and as a source of external restraints. Refinement was carried out using Refmac5 (Murshudov et al., 2011), using a coarse TLS model including only two groups, one encompassing the entire membrane domain and the other defined as the stromal ridge (PsaC, PsaD and PsaE). The use of external restraints (as well as NCS restraints) was vital to achieving reasonable statistics and ProSMART (Nicholls et al., 2012) was found to be the most effective choice in our case. Multiple rounds of refinements followed by manual building by coot were carried out. PHENIX was used in the final rounds solely for B factor and TLS refinement. The use of the higher resolution Synechocystis monomeric model resulted in a ∼1.5% reduction in R values (both working and free) and more importantly resulted in better maps. The improvement in map quality allowed us to complete the tracing of PsaK (without side chains), which is very disordered in our monomeric model and was not traced completely even in the high-resolution structure from Thermosynechococcus.
Cytochrome c553 purification
Request a detailed protocolThe soluble fraction after membrane sedimentation was used as a starting material for purification. Soluble material from 100 l culture was first clarified using 50% ammonium sulfate precipitation, than the remaining material in the supernatant was precipitated using 100% ammonium sulfate. The precipitated material was dialyzed over-night and loaded on a DEAE column equilibrated with 10 mM Tris–HCl pH 7.4. The column was eluted using a linear NaCl gradient (0–200 mM), pink fractions were collected and concentrated using a centricon (5 kDa cutoff). The concentrated pink fraction was loaded on a Superdex 75 gel filtration column. The pink protein peak coming off this column contained a single 10 kDa polypeptide according to SDS-PAGE and displayed the appropriate absorbance spectrum for cytochrome c553.
P700 reduction kinetics
Request a detailed protocolKinetic measurements were carried out using a JTS-10 spectrophotometer. Reaction mixture (for the data presented in Figure 3A,B) included 35 µg chl of purified PSI, 60 mM MES pH-6, 0.02 %DDM, 25 mM MgCl2, 50 mM NaCl, 5 mM ascorbate, 5 µM methyl viologen and cytochrome C553 (1 µM) or horse heart cytochrome c (2.5 µM) in a total volume of 1 ml. A 100 ms light pulse was applied from an orange LED source (630 nm) and three independent traces were averaged for each experiment.
Light-dependent cytochrome oxidation
Request a detailed protocolAbsorbance changes (550 nm–540 nm) were recorded using an Aminco DW-2 spectrophotometer. The components were mixed in a 1 ml quartz cuvette and they included 40 µg chl of purified PSI, 30 mM Tricine-HCl pH 8, 25 mM NaCl, 10 mM ascorbate, 10 µM methyl viologen, 5 mM horse heart cytochrome C or 2.5 mM cytochrome C553. Light was provided from a slide projector and filtered through Appropriate filters.
Data availability
-
Crystal structure of a virus like photosystem I from the cyanobacterium Synechocystis PCC 6803ID 4L6V. Publicly available at the RCSB Protein Data Bank (http://www.rcsb.org/).
-
Crystal structure of a virus like photosystem I from the cyanobacterium Synechocystis PCC 6803ID 4KT0. Publicly available at the RCSB Protein Data Bank (http://www.rcsb.org/).
References
-
PHENIX: a comprehensive Python-based system for macromolecular structure solutionActa crystallographica Section D, Biological crystallography 66:213–221.https://doi.org/10.1107/S0907444909052925
-
Bidirectional electron transfer in photosystem I: replacement of the symmetry-breaking tryptophan close to the PsaB-bound phylloquinone A1B with a glycine residue alters the redox properties of A1B and blocks forward electron transfer at cryogenic temperaturesBiochimica et Biophysica Acta 1757:1623–1633.https://doi.org/10.1016/j.bbabio.2006.07.006
-
Structure determination and improved model of plant photosystem IThe Journal of Biological Chemistry 285:3478–3486.https://doi.org/10.1074/jbc.M109.072645
-
Engine of life and big bang of evolution: a personal perspectivePhotosynthesis Research 80:137–155.https://doi.org/10.1023/B:PRES.0000030662.04618.27
-
Daddy, where did (PS)I come from?Biochimica et Biophysica Acta 1507:291–310.https://doi.org/10.1016/S0005-2728(01)00209-2
-
The origin and evolution of oxygenic photosynthesisTrends in Biochemical Sciences 23:94–97.https://doi.org/10.1016/S0968-0004(98)01186-4
-
Photosynthetic reaction center genes in green sulfur bacteria and in photosystem 1 are relatedProceedings of the National Academy of Sciences of the United States of America 89:8135–8139.https://doi.org/10.1073/pnas.89.17.8135
-
Molecular cloning and targeted mutagenesis of the gene psaF encoding subunit III of photosystem I from the cyanobacterium Synechocystis sp. PCC 6803The Journal of Biological Chemistry 266:20146–20151.
-
Light-harvesting in photosystem IPhotosynthesis Research 116:153–166.https://doi.org/10.1007/s11120-013-9838-x
-
Scaling and assessment of data qualityActa crystallographica Section D, Biological crystallography 62:72–82.https://doi.org/10.1107/S0907444905036693
-
Isolation of a psaF-deficient mutant of Chlamydomonas reinhardtii: efficient interaction of plastocyanin with the photosystem I reaction center is mediated by the PsaF subunitThe EMBO Journal 14:4976–4984.
-
Unraveling the photosystem I reaction center: a history, or the sum of many effortsPhotosynthesis Research 80:109–124.https://doi.org/10.1023/B:PRES.0000030657.88242.e1
-
The reaction center of green sulfur bacteria(1)Biochimica et biophysica acta 1507:260–277.https://doi.org/10.1016/S0005-2728(01)00200-6
-
Crystallization of intact and subunit L-deficient monomers from Synechocystis PCC 6803 photosystem IZ Naturforsch C 51:195–199.
-
XdsActa crystallographica Section D, Biological crystallography 66:125–132.https://doi.org/10.1107/S0907444909047337
-
Single core polypeptide in the reaction center of the photosynthetic bacterium Heliobacillus mobilis: structural implications and relations to other photosystemsProceedings of the National Academy of Sciences of the United States of America 90:7124–7128.https://doi.org/10.1073/pnas.90.15.7124
-
The evolution of photosystem I in light of phage-encoded reaction centresPhilosophical transactions of the Royal Society of London Series B, Biological sciences 367:3400–3405.https://doi.org/10.1098/rstb.2012.0057
-
REFMAC5 for the refinement of macromolecular crystal structuresActa crystallographica Section D, Biological crystallography 67:355–367.https://doi.org/10.1107/S0907444911001314
-
Photosystems and global effects of oxygenic photosynthesisBiochimica et Biophysica Acta 1807:856–863.https://doi.org/10.1016/j.bbabio.2010.10.011
-
The complex architecture of oxygenic photosynthesisNature Reviews Molecular Cell Biology 5:971–982.https://doi.org/10.1038/nrm1525
-
The structure of photosystem I and evolution of photosynthesisBioEssays: News and Reviews in Molecular, Cellular and Developmental Biology 27:914–922.https://doi.org/10.1002/bies.20278
-
Structure and function of photosystems I and IIAnnual Review of Plant Biology 57:521–565.https://doi.org/10.1146/annurev.arplant.57.032905.105350
-
Low-resolution refinement tools in REFMAC5Acta Crystallographica Section D, Biological Crystallography 68:404–417.https://doi.org/10.1107/S090744491105606X
-
Structural insights into energy regulation of light-harvesting complex CP29 from spinachNature Structural & Molecular Biology 18:309–315.https://doi.org/10.1038/nsmb.2008
-
Evolution of new protein topologies through multistep gene rearrangementsNature Genetics 38:168–174.https://doi.org/10.1038/ng1717
-
Marine cyanophages: tinkering with the electron transport chainThe ISME Journal 5:1568–1570.https://doi.org/10.1038/ismej.2011.43
-
Orientation resolving dipolar high-field EPR spectroscopy on disordered solids: II. Structure of Spin-Correlated Radical pairs in photosystem IThe Journal of Physical Chemistry B 117:11184–11199.https://doi.org/10.1021/jp401573z
-
Comparative metagenomics of microbial traits within oceanic viral communitiesThe ISME Journal 5:1178–1190.https://doi.org/10.1038/ismej.2011.2
-
Addition of C-terminal histidyl tags to PsaL and PsaK1 proteins of cyanobacterial photosystem IIndian Journal of Biochemistry & Biophysics 37:433–440.
-
From red to blue to far-red in Lhca4: how does the protein modulate the spectral properties of the pigments?Biochimica et Biophysica Acta 1817:711–717.https://doi.org/10.1016/j.bbabio.2012.02.030
-
Overview of the CCP4 suite and current developmentsActa Crystallographica Section D, Biological Crystallography 67:235–242.https://doi.org/10.1107/S0907444910045749
-
Function and organization of photosystem I in a cyanobacterial mutant strain that lacks PsaF and PsaJ subunitsThe Journal of Biological Chemistry 269:3205–3211.
Article and author information
Author details
Funding
European Research Council (293579-HOPSEP)
- Yuval Mazor
- Daniel Nataf
- Hila Toporik
- Nathan Nelson
Israel Science Foundation (204-10)
- Yuval Mazor
- Daniel Nataf
- Hila Toporik
- Nathan Nelson
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
Yuval Mazor would like to thank Ofer Rog for critically reading the manuscript. The authors would like to thank the ESRF, SLS and BESSYII synchrotrons for beam time and the staff scientists for excellent guide and assistance.
Copyright
© 2014, Mazor et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 2,778
- views
-
- 324
- downloads
-
- 75
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 75
- citations for umbrella DOI https://doi.org/10.7554/eLife.01496





