Small molecule-mediated refolding and activation of myosin motor function
Figures
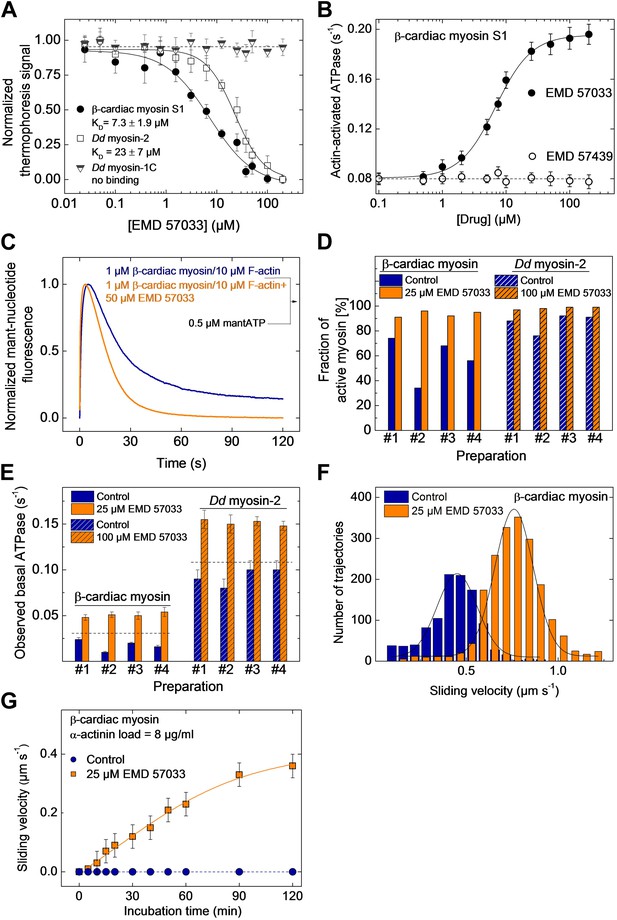
EMD 57033 binds to the myosin motor domain and activates ATPase and motor activities.
(A) Direct interaction study between fluorescently labeled β-cardiac myosin S1, Dd myosin-2 motor domain, and Dd myosin-1C motor domain and EMD 57033. The myosin concentration was kept constant at 100 nM and EMD 57033 was titrated from 10 nM to 200 µM. The normalized thermophoresis signals were plotted against the EMD 57033 concentration. KD values were obtained by fitting the data to the Hill equation. Error bars indicate SD (n = 3). (B) Dose-dependent activation of the actin-activated ATPase of β-cardiac myosin S1 by EMD 57033. Control measurements with the (−)-enantiomer EMD 57439 show no activation. Errors indicate s.d. (n = 4). (C) Single-turnover analysis of mantATP binding, hydrolysis, and product release by β-cardiac myosin in the absence (blue curve) and presence of 25 µM EMD 57033 (orange curve). (D) Fraction of active myosin heads in the absence and presence of saturating concentrations of EMD 57033 determined by active site titrations. Four β-cardiac myosin and four Dd myosin-2 preparations were compared. In the absence of EMD 57033 (blue bars) β-cardiac myosin preparations display between 34% and 74% and Dd myosin-2 preparations 76–92% activity. The fraction of active protein increased to 87–100% following the addition of EMD 57033 (25 µM for β-cardiac myosin and 100 µM for Dd myosin-2). Addition of EMD 57033 to fully denatured myosin aggregates followed by incubation for 6 hr on ice or at 20°C produced no detectable recovery of enzymatic activity. (E) Activation of basal ATPase activity by EMD 57033. The observed turnover of ATP is 1.4-fold (Dd myosin-2) and 1.5-fold (β-cardiac myosin) higher than expected for preparations of 100% active protein (dashed line). (F) EMD 57033-mediated activation of motor activity. The histograms and Gaussian fits show the distribution and average sliding velocity of actin filaments on lawns of β-cardiac myosin in the absence (blue histograms) and presence of EMD 57033 (orange histograms). Errors indicate SD (G) EMD 57033-mediated increase in force production. A constant frictional load of 8 µg/ml α-actinin was applied to stall β-cardiac myosin-based motility in the absence of EMD 57033. Filament movement is restarted after the addition of EMD 57033. Errors indicate SD.
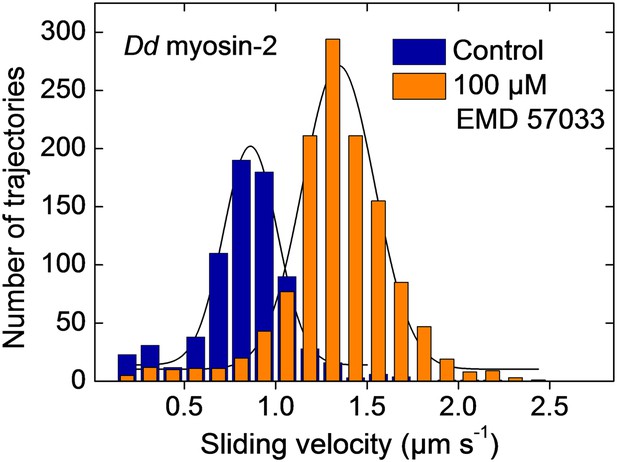
EMD 57033-mediated activation of motor activity for a recombinant Dd myosin-2 motor domain construct.
The histograms and Gaussian fits show the distribution and average sliding velocity of actin filaments on lawns of Dd myosin-2 motor (blue histograms). In the presence of EMD 57033 (100 µM, orange histograms) the sliding velocity is increased from 0.86 ± 0.15 µm s−1 to 1.35 ± 0.2 µm s−1. Errors indicate SD.
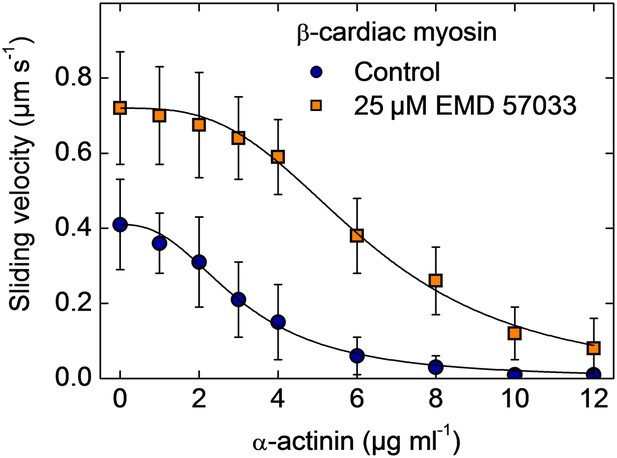
Frictional loading experiments.
Faster filament movment is observed in the presence of EMD 57033 and higher concentrations of α-actinin are required to stall filament movement on surfaces decorated with β-cardiac myosin. Errors indicate SD.
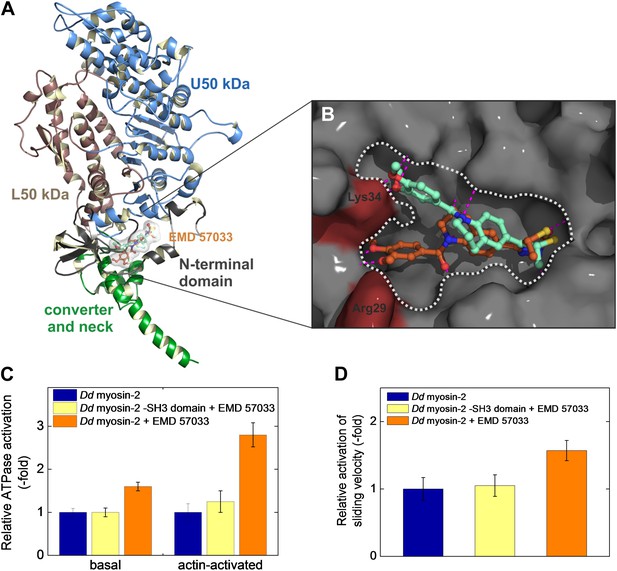
Localization of the EMD 57033 binding pocket.
(A) Predicted binding of EMD 57033 near the base of the lever arm as obtained by molecular docking. The overview of the Hs β-cardiac myosin head domain includes residues 1–800. Two slightly different clusters of binding poses were identified for EMD 57033 with comparable binding affinities. (B) Close-up of the allosteric binding pocket shown in surface representation. The Y shape of the pocket is outlined and the two residues—Arg29 and Lys34—that were allowed full conformational flexibility during docking are colored red. For clarity only the best poses for the two identified clusters of binding modes are shown. Hydrogen bonds of EMD 57033 to the motor protein are shown in magenta. (C) The SH3-like subdomain is required for EMD 57033 binding and myosin activation. A truncated Dd myosin-2 motor domain construct without SH3-like β-barrel shows no significant activation in the presence of 100 μM EMD 57033. Errors indicate SD (n = 4). (D) In contrast to the wild-type Dd myosin-2 motor domain construct, no actin filament sliding velocity enhancement was observed upon addition of 100 µM EMD 57033 to the construct with truncated N-terminal SH3-like β-barrel. Errors indicate SD.
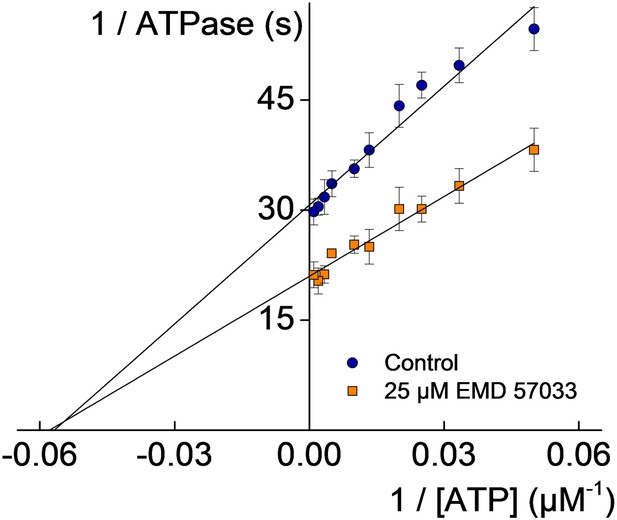
Kinetic analysis of the effect of EMD 57033 binding using the Lineweaver–Burk plot.
Rates measured in the absence (blue circles) and in the presence of 25 µM EMD 57033 (orange rectangles). KM,ATP remains unaffected, whereas kcat is 1.5-fold increased. Errors indicate SD.
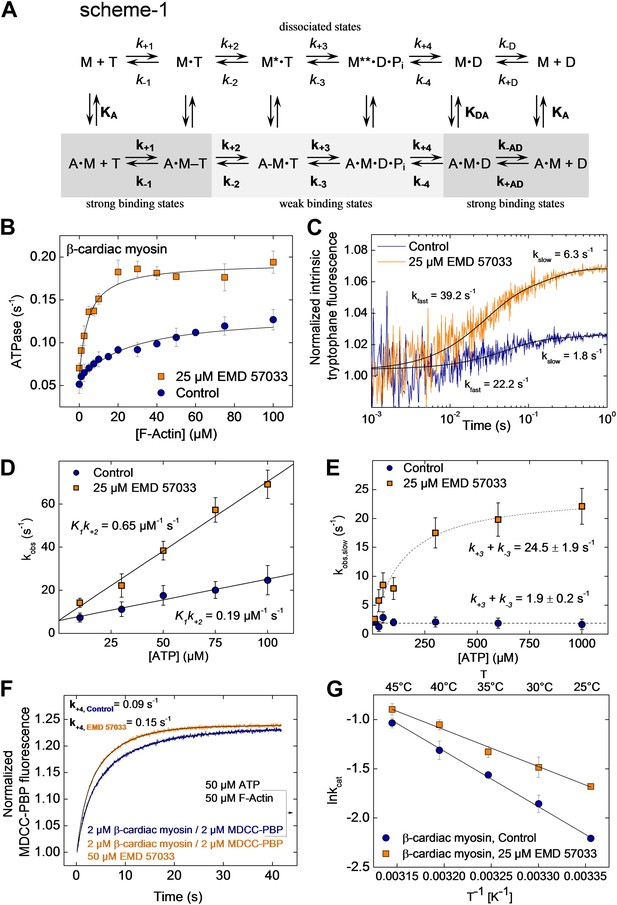
EMD 57033-mediated changes in the interaction with nucleotides and F-actin.
(A) Kinetic reaction scheme of the actomyosin ATPase cycle. ‘A’ refers to actin, ‘M’ to myosin, ‘T’ to ATP, and ‘D’ refers to ADP. Rate constants are referred to as k+n and k−n and are assigned to the corresponding forward and reverse reactions. An additional notation is used that distinguishes between the constants in the absence and presence of actin by italic type (k+1, K1) and bold (k+1, K1), respectively; subscript A refers to actin (KA) and subscript D (KD) refers to ADP (B) EMD 57033-mediated increase in the actin-dependence of ATP-turnover. In the presence of 25 µM EMD 57033, the plateau value representing kcat is 1.6-fold increased and the apparent actin affinity KM(Actin), indicated by the half-maximal activation of ATPase activity, is 10-fold increased (values are given in Table 2). Errors indicate SD (n = 5). (C) EMD 57033-mediated increase in the rate of ATP binding and hydrolysis. The fast phase of the biphasic transients correspond to ATP binding, the slow phase can be attributed to ATP hydrolysis. (D) EMD 57033-mediated changes in the rate of ATP binding. The observed rates for the fast phase show a linear dependence on ATP concentration. Differences in the slopes indicate a threefold increase of the second order rate constants for ATP binding in the presence of EMD 57033. Errors indicate SD. (E) EMD 57033 mediates a 10-fold increase in the rate of ATP hydrolysis. The plateau values from a hyperbolic fit of the observed slow components of the fluorescence transients plotted against ATP concentrations define the rate of ATP hydrolysis. Errors indicate SD. (F) The rate of phosphate release increased 1.7-fold in the presence of EMD 57033. Phosphate release from β-cardiac myosin was observed following rapid mixing with excess ATP and F-actin in the presence of the fluorescent phosphate-binding protein MDCC-PBP. Errors indicate SD. (n = 6 from three different protein preparations). (G) Arrhenius analysis of the temperature dependence of β-cardiac myosin ATPase activity. The activation energy for the reaction is 1.5-fold reduced in the presence of EMD 57033. Errors indicate SD. (n = 3).
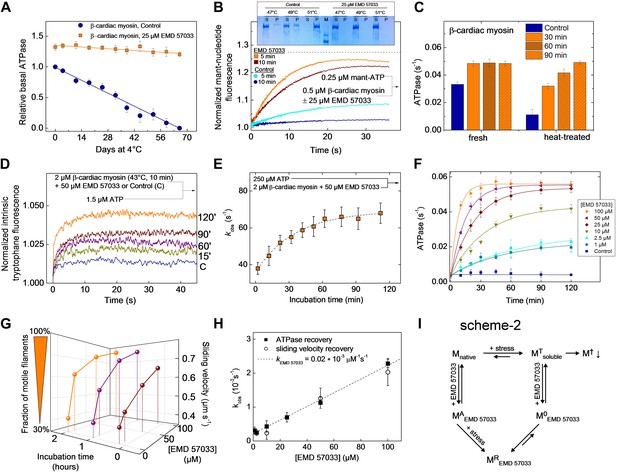
EMD 57033 acts as a pharmacological chaperone.
(A) The presence of 25 µM EMD 57033 extends the shelf life of Hs β-cardiac myosin at 4°C. Errors indicate SD (n = 3). (B) EMD 57033 renders β-cardiac myosin more heat-stable. The SDS-PAGE gel shows supernatants (S) and pellets (P) after 10-min incubation of β-cardiac myosin at the indicated temperatures. In the presence of EMD 57033 β-cardiac myosin remained in the supernatant over the entire temperature range tested. Incubation for 10 min at 49°C leads to the almost complete loss of catalytic activity. The ability to bind mantATP is gradually recovered following the addition of 25 µM EMD 57033. The dotted line represents the fluorescence amplitude for mantATP binding to native β-cardiac myosin. (C) Rescue and activation of heat-treated β-cardiac myosin basal ATPase activity by EMD 57033. The orange columns indicate changes observed in the presence of EMD 57033. Errors indicate SD (n = 3). (D) Time-dependent recovery of the capacity to bind nucleotide monitored by ATP-induced changes in intrinsic tryptophan fluorescence. Refolding was initiated by the addition of 50 µM EMD 57033. Transients obtained after rapid mixing with ATP were measured at the indicated times following the addition of EMD 57033. (E) The observed rate constants for ATP binding to heat-treated β-cardiac display a hyperbolic dependence upon the incubation with EMD 57033. Errors indicate s.d. (F) Effect of EMD 57033 concentration on the time-dependent recovery of heat-treated β-cardiac myosin ATPase activity. Errors indicate SD (n = 3). (G) Time and EMD 57033 concentration dependence of the recovery of β-cardiac myosin motor activity. (H) Determination of the second order rate constant for the EMD 57033-mediated refolding reaction. The observed rate constants for the recovery of ATPase activity and motility were extracted from the data shown in Figure 4F,G. The slope of a linear fit to the data defines krescue as 0.02 × 10−3 μM−1s−1. The y-intercept gives a first order dissociation rate constant for EMD 57033 of 0.23 × 10−3 s-1 (I) Proposed model for the mode of action of EMD 57033. T Conformationally trapped soluble aggregates, nucleotide binding incompetent; R Rescued, hydrolysis and motility competent; 0 Compromised, hydrolysis incompetent, nucleotide binding competent; A Activated; † Insoluble protein aggregates (irreversible); ↓ Precipitation.
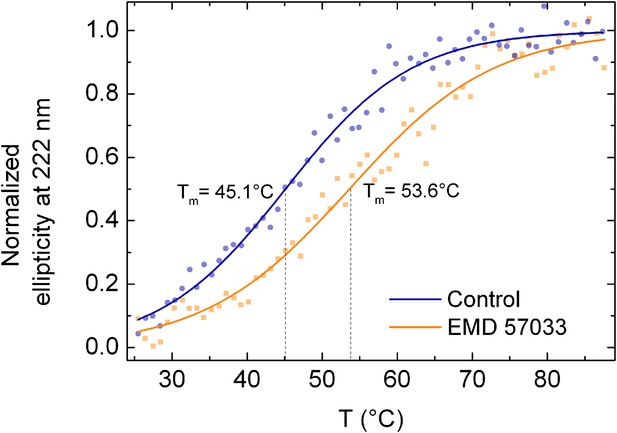
Melting curves of the β-cardiac myosin motor domain as determined by circular dichroism spectroscopy at 222 nm (c = 0.3 mg/ml).
Circular Dichroism was followed along a temperature gradient from 25°C to 90°C in the absence and presence of 100 µM EMD 57033. Data curves are averaged from 3 individual measurements, normalized and fitted according to the model of Boltzmann. [0] displays the folded state whereas [1] is attributed to the unfolded state. The melting temperatures are 45.1 ± 2.2°C in the absence of EMD 57033 and 53.6 ± 1.9°C in its presence, respectively.
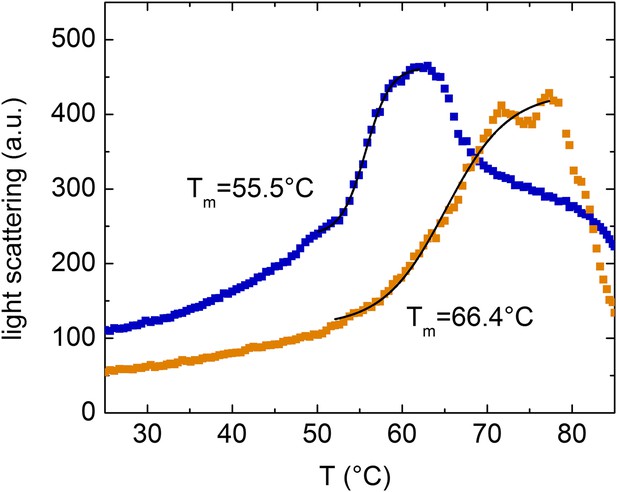
Melting curves of the complex formed by the β-cardiac myosin motor domain with F-actin followed by the change in light-scattering signal.
Light scattering was followed along a temperature gradient from 25°C to 90°C in the absence and presence of 100 µM EMD 57033. The mid-points of the observed transitions correspond to approximately 55.5°C in the absence and 66.4°C in its presence of EMD 57033.

Effects of EMD 57033 on neonatal rat cardiomyocytes (NRCM) in vitro.
(A) Cell size and organization of the sarcomere in representative NRCM (DMSO control) at 37°C. (B) Representative NRCM after 24 hr incubation with EMD 57033. Cardiomyocytes were immunostained for α-actinin (green) and DAPI was used for nuclear staining. (C) Bar graph depicting cumulative measurements of cell size of NRCM after 24 hr incubation with EMD 57033 or DMSO at 37°C or 42°C. NRCM display a comparable increase in cell size in response to EMD 57033 stimulation independent of the incubation temperature. (D) Bar graph showing expression of α-cardiac MHC mRNA normalized to b2m in NRCM after 24 hr treatment with EMD 57033 or DMSO. EMD 57033 does not alter α-cardiac MHC mRNA expression compared to DMSO controls. Incubation at 42°C significantly reduces expression of α-cardiac MHC mRNA in NRCM compared to cells that were incubated at 37°C. The decrease in α-cardiac MHC mRNA upon induction of hyperthermia was not affected by EMD 57033 treatment. Errors indicate SD (n = 3). (E) Bar graph summarizing mRNA expression of β-cardiac MHC normalized to b2m in EMD 57033 or DMSO-treated NRCM after 24-hr incubation at 37°C or 42°C. Treatment with EMD 57033 significantly reduces β-cardiac MHC expression at 37°C compared to DMSO controls. Incubation at 42°C for 24 hr results in downregulation of β-cardiac MHC mRNA. Downregulation is significantly more pronounced in EMD 57033-treated cells. Errors indicate SD (n = 3). (F) Representative immunoblot depicting expression of total cardiac MHC protein content in NRCM treated with EMD 57033 or DMSO. EMD 57033 treatment does not significantly alter MHC protein levels at 37°C or 42°C and MHC protein content is comparable in NRCM incubated at 37°C or 42°C. (G) Bar graph depicting mRNA expression of ANP normalized to b2m in NRCM treated with EMD 57033 or DMSO for 24 hr at 37°C or 42°C. EMD 57033 treatment does not affect expression of ANP mRNA at 37°C. Incubation of NRCM at 42°C induces mRNA expression of ANP in DMSO-treated control cells, which is completely suppressed by treatment with EMD 57033. Errors indicate SD (n = 3). NRCM were treated with EMD 57033 (10 µM) or the solvent DMSO (1 µl/ ml) alone for 24 hr in serum-free conditions. Significances are coded as follows: *p<0.05, **p<0.01 und ***p<0.001 for the DMSO control vs EMD 57033 at a specific temperature; #p<0.05, ##p<0.01 und ###p<0.001 for a specific group at different temperatures.
Videos
Video showing fluorescently labeled actin filaments attached to a lawn of catalytically inactive β-cardiac myosin. (MT state, Figure 4I).
https://doi.org/10.7554/eLife.01603.014Reactivation of β-cardiac myosin by the addition of EMD 57033.
Video showing motile actin filaments on the same lawn of β-cardiac myosin three hours after the addition of 10 µM EMD 57033 to the flow cell (MR state, Figure 4I).
Tables
Interaction of EMD 57033 with myosin isoforms
| Myosin construct | AC50 ATPase | Normalized maximal ATPase activation (basal) | Normalized maximal ATPase activation (with 30 µM actin) | Binding affinity (MST) |
|---|---|---|---|---|
| β-Cardiac myosin-2 (S1, full-length) | 7.0 µM | 1.5 | 2.5 | 7.3 µM |
| Skeletal muscle myosin-2 (HMM) | 15.1 µM | 1.6 | 2.2 | – |
| Dd myosin-2 motor domain | 25.8 µM | 1.4 | 2.8 | 23.0 µM |
| Dd myosin-5b motor domain | 35.4 µM | 1.4 | 1.6 | – |
| Dd myosin-1B, -1C, -1D, -1E motor domains | n.a. | no effect | no binding (myosin-1C/-1D/-1E) | |
| Dd myosin-2 ΔSH3 22 (lacks residues 33–79) | n.a. | no effect | n.a. | |
EMD 57033-mediated changes in the kinetic behavior of β-cardiac myosin
| Control | 25 µM EMD 57033 | ∼Change (−fold) | |
|---|---|---|---|
| KM(Actin) | 36. 8 ± 2.67 µM | 3. 6 ± 0.8 µM | 10 |
| kcat | 0. 12 ± 0.02 s−1 | 0.19 ± 0.02 s−1 | 1.6 |
| kcat/KM(Actin)* | 0.00326 µM−1 s−1 | 0.0528 µMv1 s−1 | 16 |
| kcat/KM(Actin)† | 0.00193 µM−1 s−1 | 0.0304 µM−1 s−1 | 15.8 |
| K1k+2 | 0.19 ± 0.02 µM−1s−1 | 0.65 ± 0.04 µM−1s−1 | 3.4 |
| k+2 | 46 ± 3 s−1 | 174 ± 7 s−1 | 3.8 |
| k+3 + k−3 | 1.9 ± 0.2 s−1 | 24.5 ± 1.9 s−1 | 12.9 |
| k+4‡ | 0.09 ± 0.01 s−1 | 0.15 ± 0.01 s−1 | 1.7 |
| k−D | 0.15 ± 0.03 s−1 | 0.16 ± 0.04 s−1 | n.a. |
| k−AD | 25.5 ± 3 s−1 | 26.9 ± 4 s−1 | n.a. |
| Ea | 47 ± 4 kJ mol−1 | 31 ± 3 kJ mol−1 | 1.5 |
-
*
The apparent second order rate constant for actin binding (kcat/KM(Actin)) was obtained from the calculated ratio of both values.
-
†
kcat/KM(Actin) was obtained from the initial slope of the steady-state ATPase activity vs the F-actin plot.
-
‡
Measured at F-Actin concentration of 25 µM and 25 µM ATP.
Primer sequences and PCR conditions used for realtime PCR
| Primer | Primer-Sequence | Annealing temperature (°C) |
|---|---|---|
| r-ANP forward | 5′-GCCGGTAGAAGATGAGGTCA-3′ | 60 |
| r-ANP reverse | 5′-GGGCTCCAATCCTGTCAATC-3′ | 60 |
| r-aMHC forward | 5′-GGAAGAGCGAGCGGCGCATCAAGG-3′ | 55 |
| r-aMHC reverse | 5′-GTCTGCTGGAGAGGTTATTCTCG-3′ | 55 |
| r-bMHC forward | 5′-CAAGTTCCGCAAGGTGC-3′ | 55 |
| r-bMHC reverse | 5′-AAATTGCTTTATTGTGTTTCT-3′ | 55 |
| r-b2m forward | 5′-CATGGCTCGCTCGGTGACC-3′ | 60 |
| r-b2m reverse | 5′-AATGTGAGGCGGGTGGAACTG-3′ | 60 |





