Delivery of endocytosed proteins to the cell–division plane requires change of pathway from recycling to secretion
Figures
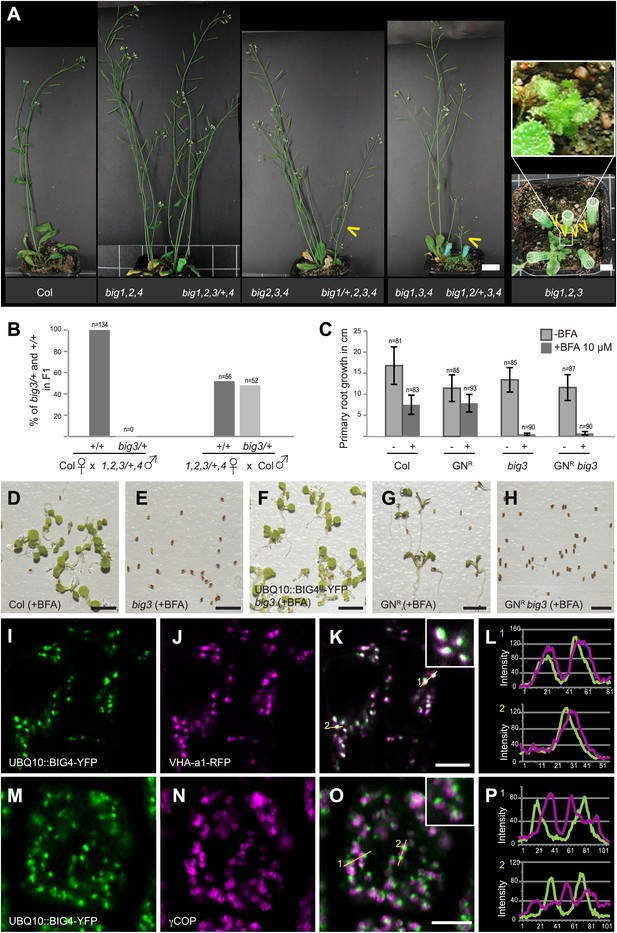
BIG1 – BIG4 act redundantly at TGN and are involved in several physiological processes.
(A) big1,2,4 (big1 big2 big4), big2,3,4 (big2 big3 big4), big1,3,4 (big1 big3 big4) and big1,2,3/+,4 (big1 big2 big3/BIG3 big4) mutant plants without obvious phenotype but big1/+,2,3,4 (big1/BIG1 big2 big3 big4), big1,2/+,3,4 (big1 big2/BIG2 big3 big4) and big1,2,3 (big1 big2 big3) were dwarfed (yellow arrowheads). Scale bar, 2 cm. (B) F1 of reciprocal crosses between wild-type (Col) and big1 big2 big3/BIG3 big4 (1,2,3/+,4) mutants: 0% or 48% big3 heterozygous seedlings derived from mutant male or female gamete, respectively. (C) BFA inhibited primary root growth of big3 mutant seedlings with or without BFA-resistant GNOM (GNR big3). Numbers of analysed seedlings are indicated (B and C). (D-H) BFA treatment did not prevent seed germination in wild-type (Col; D) and BFA-resistant GN (GNR; G) but did so in big3 mutants without (E) or with BFA-resistant GNOM (GNR big3; H). This defect was suppressed by BFA-resistant BIG4 (UBQ10::BIG4R-YFP big3; F). Scale bar, 5 mm. (I-L) Live imaging of BIG4-YFP (I) and TGN marker VHA-a1-RFP (J) revealed co- localization (K; L, intensity–line profile). (M–P) Immunolocalization of BIG4 (UBQ10::BIG4-YFP; M) and Golgi-marker γCOP (N) indicated no co-localization (O; P, intensity–line profile). (I–K, M–O) Scale bar, 5 μm.
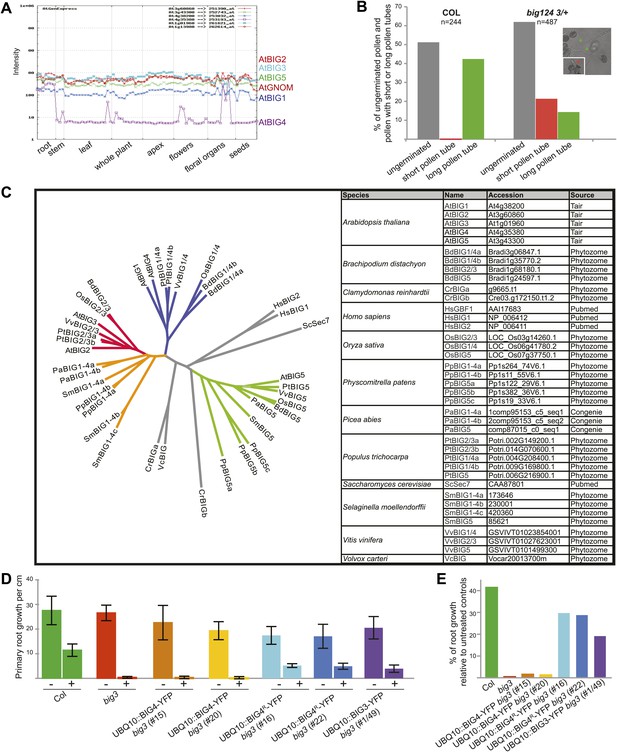
Expression and phylogeny of BIG ARF-GEFs.
(A) Expression profiles of BIG1–BIG5 and GNOM. Data from AtGenExpress (Schmid et al., 2005) (http://jsp.weigelworld.org). Note preferential expression of BIG4 in roots and floral organs. (B) Analysis of in vitro pollen tube growth of pollen from wild-type (Col) and big124 3/+ plants. Note approximately 50% of germinated pollen from mutant plants produced short tubes (red column); asterisks: red, short; green, long pollen tubes). (C) Phylogenetic tree (ClustalW) of BIG ARF-GEFs from flowering plants (dicots Arabidopsis [At], poplar [Pt] and grapevine [Vv]; monocots rice [Os] and Brachypodium [Bd]), gymnosperm Picea abies (Pa), lower plants (lycopod Selaginella [Sm], moss Physcomitrella [Pp], and algae Chlamydomonas [Cr] and Volvox [Vc]), and outgroups (human [Hs], Saccharomyces cerevisiae [Sc]) (grey). Three distinct subclades (BIG1/4 [blue], BIG2/3 [red] and BIG5 [green]) are present in angiosperms. However, only BIG5 is distinct in all plant species whereas lower plants have a single subclade BIG1-4 corresponding to the two subclades BIG1/4 and BIG2/3 in angiosperms (orange). Accession numbers and source of data are listed in the table.(D and E) BFA-resistant BIG4 and BIG3 expressed from the Ubiquitin10-promoter (UBQ10::BIG4R/BIG3-YFP) can partially rescue the BFA-inhibited primary root growth of big3 mutants.(E) Percentage of root growth of BFA-treated seedlings shown in (D) relative to untreated controls.
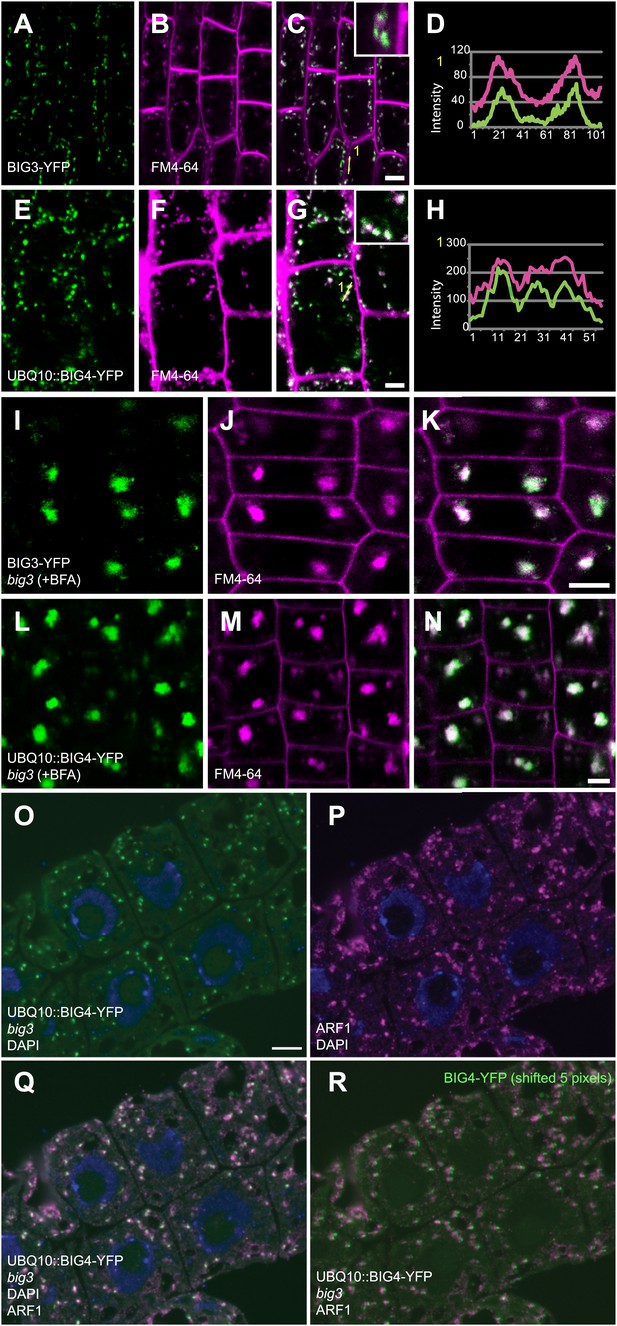
BIG3 and BIG4 localize at the TGN.
(A–N) Live imaging of YFP-tagged BIG3 (A and I), expressed from its own promoter, and BIG4 (E and L), expressed from the UBQ10 promoter, in seedling roots counterstained with endocytic tracer FM4-64 (B, F, J, M). Both BIG3 and BIG4 co-localized with FM4-64, which visualized the TGN after 10 min of incubation (C, G; D, H, intensity profiles of lines numbered in C, G). (I–N) After BFA treatment, both BIG3 and BIG4 co-localized with FM4-64 in BFA compartments (K and N). Scale bars, 5 μm. (O–R) Co-localization of BIG4-YFP fluorescence (O) with immunofluorescence labeling of the TGN marker ARF1 (P) in 350 nm thin cryosections (= high axial resolution) revealed by overlay (Q) and image frames shifted by 5 pixels (R). Blue, DAPI-stained nuclei. Scale bar (O), 10 μm.
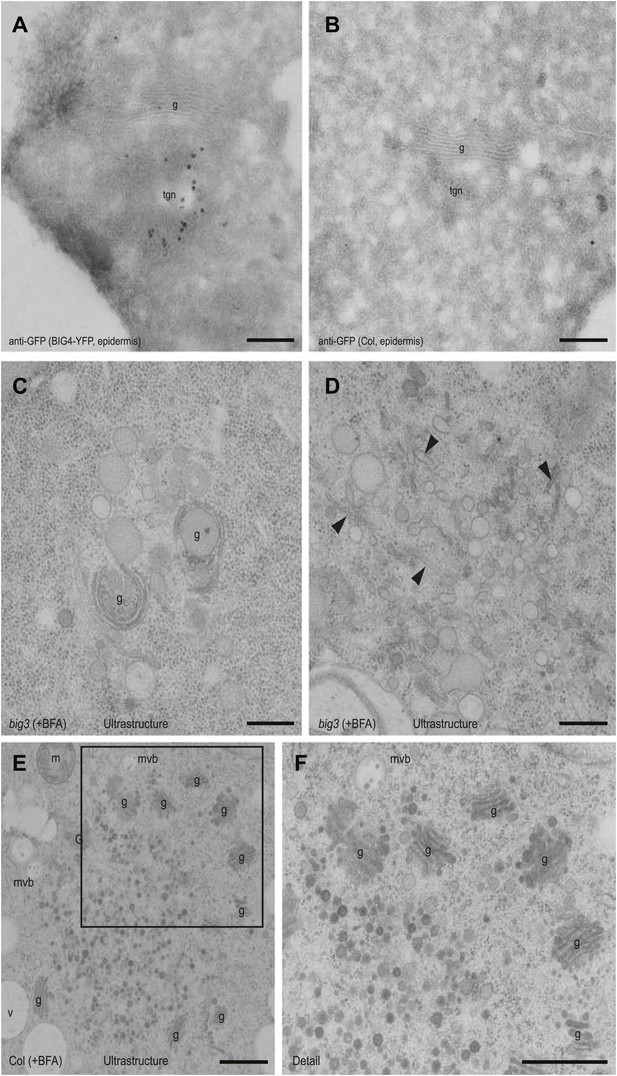
Ultrastructural localization of BIG4-YFP and ultrastructural abnormalities in BFA-treated big3 mutant seedling root cells.
(A and B) Anti-GFP immunogold labeling of YFP-tagged BIG4 in root epidermal cells of UBQ10::YFP-BIG4 transgenic (A) and Col-0 wild-type control (B) seedlings (thawed cryosection labeling). (C–F) Ultrastructural TEM analysis of BFA-treated big3 (C and D) and Col-0 wild-type (E and F) root epidermal cells. Note Golgi stacks are abnormally shaped (C and G) or reduced (D, arrowheads) near endosomal BFA aggregates (C and D), in contrast to well-formed Golgi stacks in wild-type (E and F). (F) Higher magnification of boxed area in (E). g, Golgi stack; m, mitochondrion; mvb, multivesicular body; tgn, trans-Golgi network; v, vacuole. Scale bars, 500 nm.
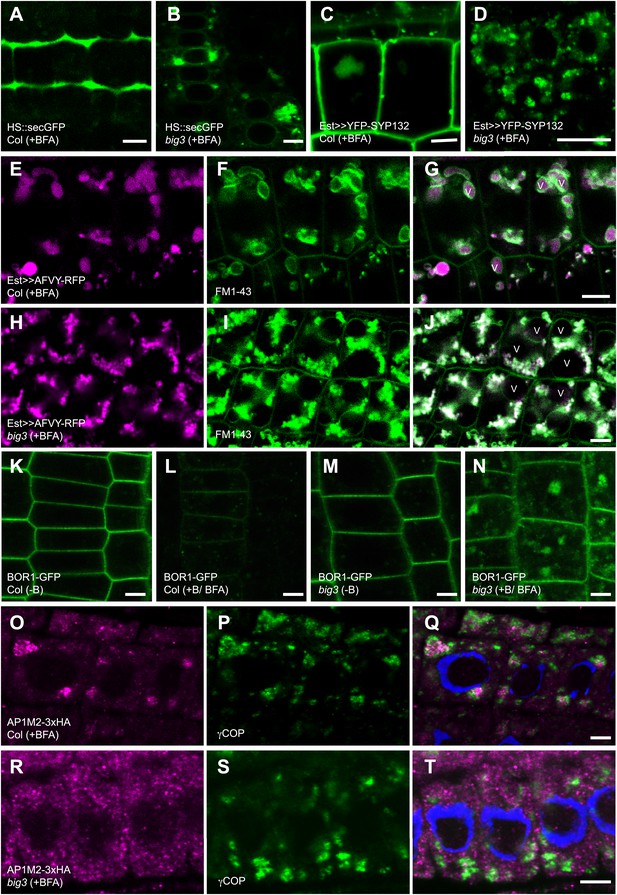
BIG1 – BIG4 regulate secretory and vacuolar trafficking by recruiting AP-1 adaptor complex.
(A and B) BFA inhibited secretion of heat shock (HS)-induced secGFP in big3 mutants (B) but not in wild-type (Col; A). (C and D) BFA inhibited trafficking of estradiol (Est)-induced YFP-SYP132 to the plasma membrane in big3 mutants (D) but not in wild-type (Col; C). (E–J) BFA inhibited trafficking of soluble cargo AFVY-RFP to the vacuole (v), labeled by FM1-43 (F and I), in big3 mutants (H–J) but not in wild-type (Col, E–G). (K–N) Live imaging of BOR1-GFP localization. Without boron (−B), BOR1-GFP localized at the plasma membrane in wild-type (K) and big3 mutants (M). After BFA and boron treatment (+B), BOR1-GFP was degraded in the vacuole of wild-type (L) but accumulated in BFA compartments of big3 mutants (N). (O–T) Immunostaining of 3xHA-tagged muB2 subunit of AP-1 complex (AP1M2; O, R) and COPI subunit γCOP (P and S) in BFA-treated seedlings. AP1M2 accumulated in BFA compartments surrounded by γCOP in wild-type (Col; Q). In big3 mutants, γCOP was still recruited to Golgi membranes whereas AP1M2 was cytosolic (R–T). Blue, DAPI-stained nuclei. Scale bars, 5 µm.
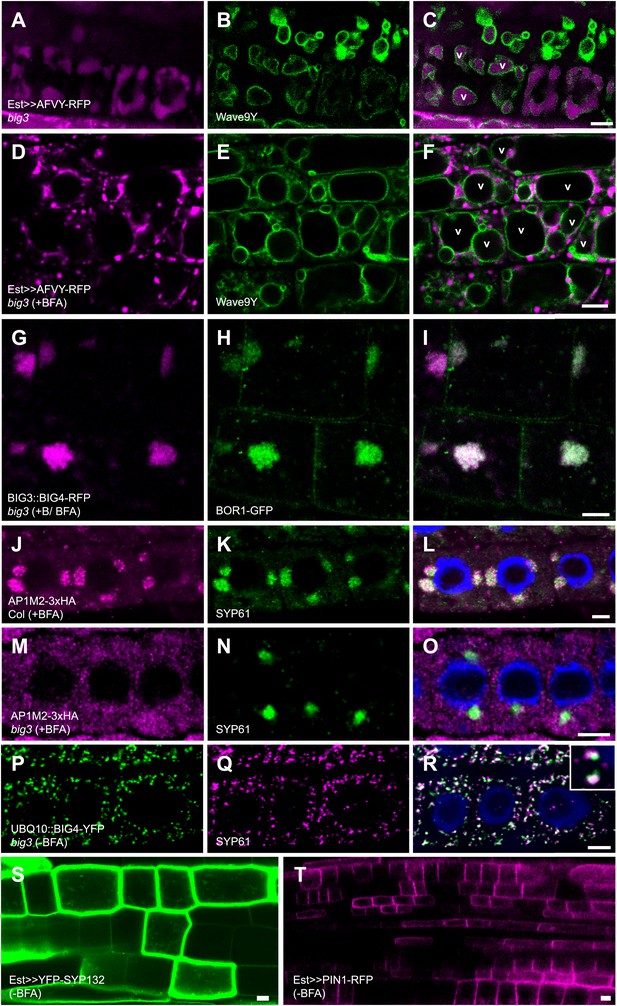
BIG1 – BIG4 regulate trafficking of secretory and vacuolar cargo by recruiting AP-1 complex.
(A–F) Live imaging of vacuolar cargo AFVY-RFP (A and D) and vacuolar membrane marker Wave9Y (Wave9-YFP/YFP-VAMP711; B, E; Geldner et al., 2009) in root cells of big3 mutant seedlings. (C and F) Overlays. Traffic of AFVY-RFP to the Wave9-labeled vacuole (v; A–C) was blocked by BFA treatment, with AFVY-RFP accumulating in small compartments distinct from vacuoles (D–F). (G–I) Live imaging of BOR1-GFP in boron (+B) and BFA-treated big3 mutants expressing BIG3-promoter driven BIG4-RFP (BIG3::BIG4-RFP). BOR1-GFP was not transported to the vacuole but co-localized with BFA-sensitive BIG4 in BFA compartments. (J–O) Immunolocalization of HA-tagged AP1M2 and TGN marker SYP61 in BFA-treated seedling roots. AP1M2 co-localized with SYP61 in BFA compartments in wild-type (J–L) but was cytosolic in big3, in contrast to TGN-associated SYP61 (M–O). (P–R) Immunostaining of UBQ10 promoter-driven BIG4-YFP and SYP61 in untreated big3 mutant seedling roots. BIG4 (P) co-localized with SYP61 (Q) at TGN (R). (S and T) YFP-SYP132 (S) and PIN1-RFP (T) expressed from the estradiol-inducible promoter localize at the plasma membrane in an unpolar or polar fashion, respectively. Scale bars, 5 μm.
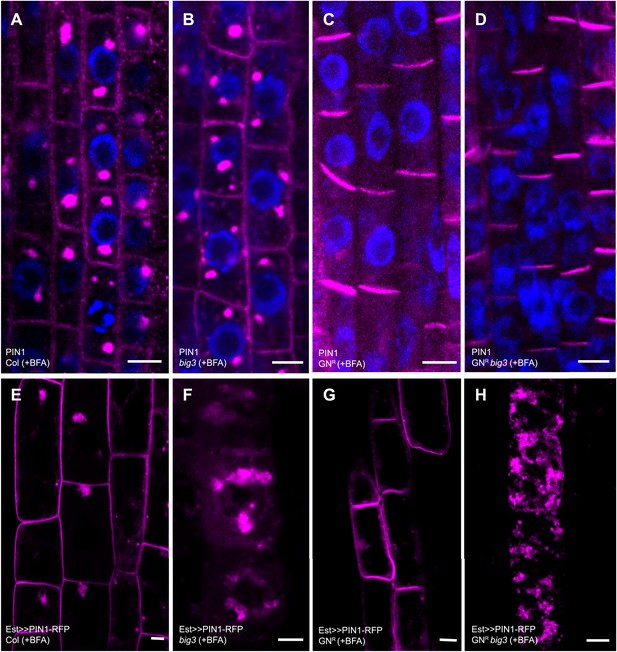
Secretion and recycling to the plasma membrane are regulated by different ARF-GEFs.
(A–D) PIN1 localization in interphase cells of BFA-treated seedlings; apolar at the plasma membrane (PM) and in BFA compartments in wild-type (Col; A) and big3 mutants (B); at the basal PM in BFA-resistant GN in wild-type (GNR, C) or big3 mutant background (GNR big3, D). Blue, DAPI-stained nuclei. (E–H) After BFA treatment, estradiol (Est)-induced PIN1-RFP was trafficked to the PM in wild-type (E) and BFA-resistant GN seedlings (GNR, G) but not in big3 mutants without (F) or with expression of BFA-resistant GN (GNR big3; H). Scale bars, 5 µm.
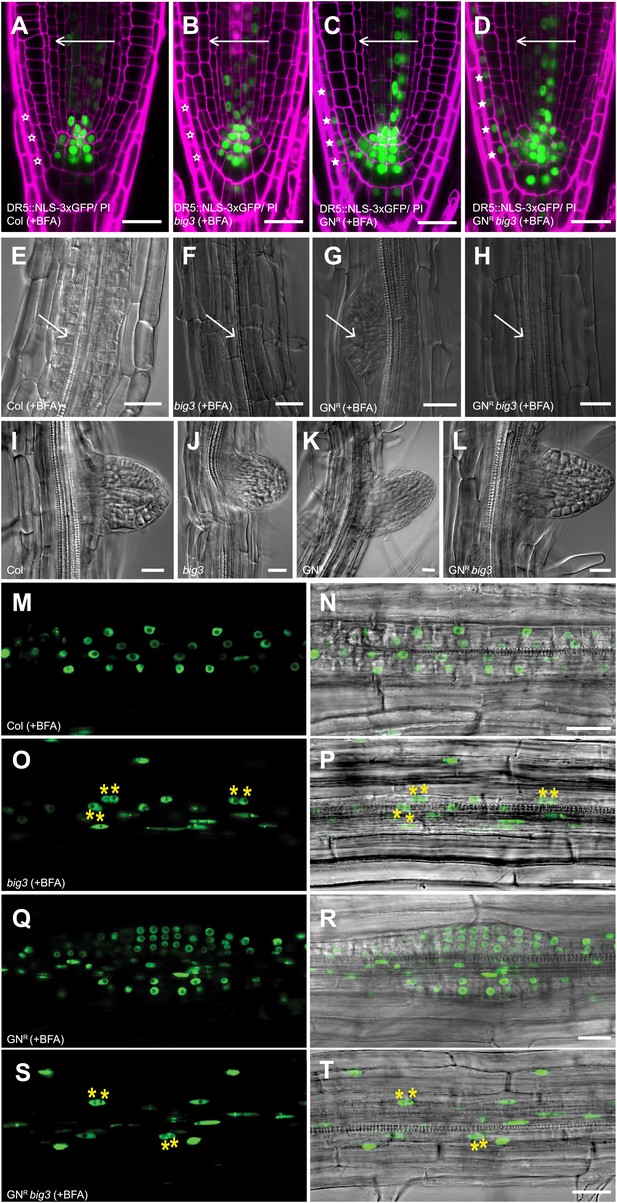
BIG1-4 in response to auxin application.
(A–D) Visualization of auxin distribution by DR5::NLS-3xGFP (green) in BFA-treated seedlings after gravistimulation. Arrows, gravity vector. Cell walls were stained by propidium iodide (PI; magenta). Wild-type (A) and big3 mutant seedling roots (B) did not respond to gravity (open asterisks), in contrast to BFA-resistant GN either in wild-type (GNR, C) or big3 mutant background (GNR big3, D). Asterisks, auxin response in epidermal cell layer on lower side (C and D). (E–H) NAA and BFA treatment led to proliferation of pericycle cells (arrows) in wild-type (E) but not big3 mutants without (F) or with BFA-resistant GN (H). Normal lateral root primordia only formed in BFA-resistant GN (GNR, G). Scale bars, 25 µm. (I–L) Bright-field microcopy of developing lateral root primordia in NAA-treated seedlings; genotypes: wild-type (Col; I), big3 (J), BFA-resistant GN (GNR; K) and BFA-resistant GN in big3 mutant background (GNR big3; L). (M–T) Live imaging of DR5::NLS-3xGFP of seedling roots after NAA and BFA treatment. DR5::NLS-3xGFP signals (left panels M, O, Q, S) overlaid with Nomarski images (right panels N, P, R, T). Pericycle cells proliferated in wild-type (M and N) but became binucleate (asterisks) in big3 (O and P) and GNR big3 (S and T) mutants. Normal lateral root primordia were only formed in BFA-resistant GN (GNR; Q, R) mutant. Scale bars, 25 µm.
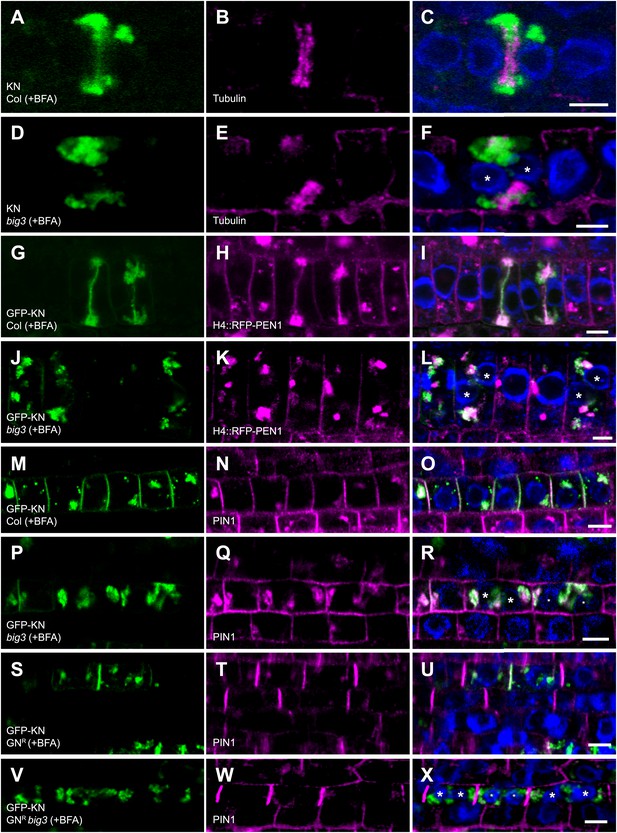
Trafficking to the plane of cell division is mediated by BIG1 – BIG4.
(A–F) Immunolocalization of KNOLLE (KN; A, D) and tubulin (B and E) in cytokinetic root cells of BFA-treated seedlings (50 µM for 3 hr). (A–C) KN was located at the cell plate (A) flanked by tubulin-positive phragmoplast (B) in wild-type. (D–F) In big3 mutants, KN accumulated in BFA compartments separated from tubulin-positive phragmoplast, resulting in a binucleate cell. (G–L) Co-localization of GFP-tagged KN and endocytosed RFP-PEN1 (H4::RFP-PEN1) in BFA-treated seedlings. KN and PEN1 co-localized at the cell plate and in BFA compartments of wild-type (G–I) but only in BFA compartments in big3 mutants (J–L). (M–X) Immunostaining of GFP-KN and PIN1 in cytokinetic root cells of BFA-treated seedlings. (M–R) PIN1 localized apolarly at the plasma membrane (PM) and co-localized with KN in BFA compartments and at the cell plate in wild-type (M–O) but only in BFA-compartments in big3 mutants (P–R). (S–U) In GNR, PIN1 localized polarly at the plasma membrane (T) and co-localized with KN (S) at the cell plate (U). (V–X) Although PIN1 localized polarly at the PM (W) in GNR big3, neither PIN1 (W) nor KN (V) was located at the cell plate. Blue, DAPI-stained nuclei. Asterisks label nuclei of binucleate cells (F, L, R, X). Scale bars, 5 µm.
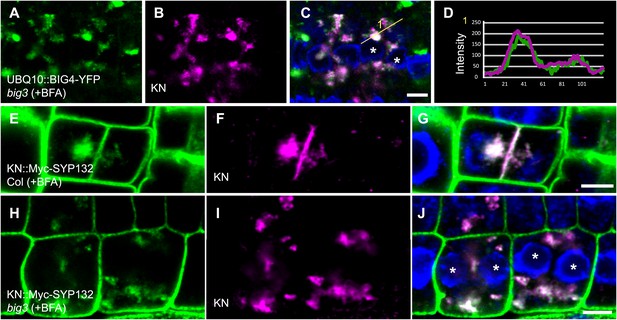
BIG4 and cargo proteins trapped in BFA compartments of dividing cells in BFA-treated big3 mutant seedlings.
(A–C) Immunostaining of UBQ10 promoter-driven BIG4-YFP and KN in BFA-treated big3 mutant seedlings. BFA-sensitive YFP-tagged BIG4 (A) co-localized with its cargo KN (B) in BFA aggregates. (C) Overlay. (D) Intensity profile of line numbered in (C) indicated overlapping signals. (E–J) Immunostaining of Myc-tagged SYP132 expressed from the KN promoter (KN::Myc-SYP132; E, H) and KN (F and I) in BFA-treated cytokinetic cells of wild-type and big3 mutant seedlings. (E–G) SYP132 localized at the plasma membrane and KN-labeled cell plate in wild-type whereas both SYP132 and KN were trapped in BFA compartments in big3 mutant (H–J). Asterisks, binucleate cells (C and J). Scale bars, 5 μm.
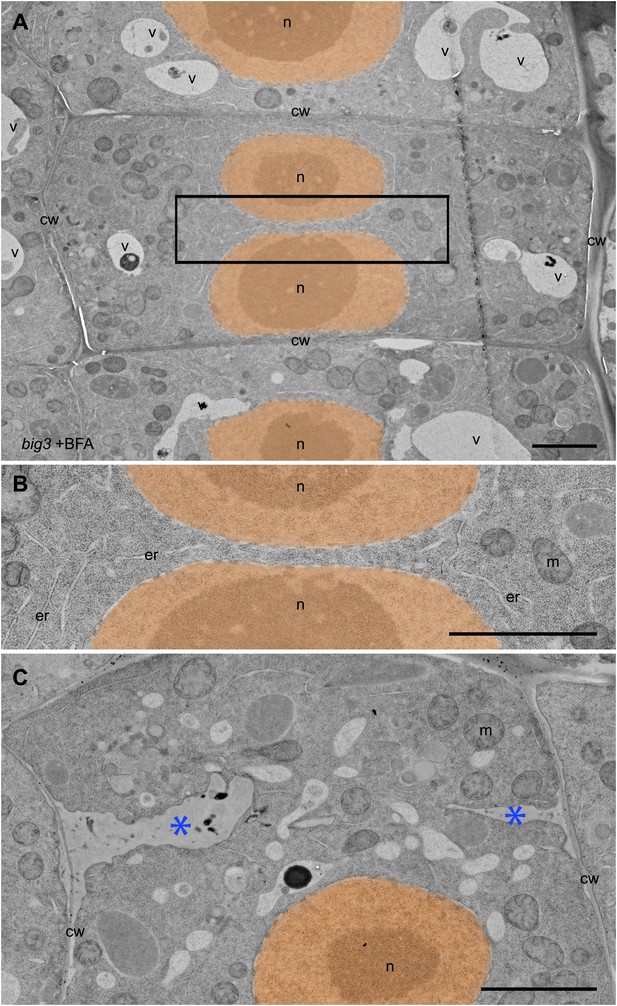
Ultrastructural appearance of cryofixed, freeze-substituted and resin-embedded big3 seedling root tips treated with BFA.
(A–C) Binucleate cells in big3 seedling roots treated with BFA. (A) Overview of ultrastructural TEM analysis. (B) Higher magnification of boxed area in (A). Note absence of cell-wall remnants or membrane vesicles between the daughter nuclei (B). (C) Another cell showing cell wall remnants (stubs, blue asterisks). Nuclei (n) have been false-colored; second nucleus in (C) is in a different focal plane. cw, cell wall; er, endoplasmic reticulum; m, mitochondrion; n, nucleus; v, vacuole. Scale bars, 2.5 μm.
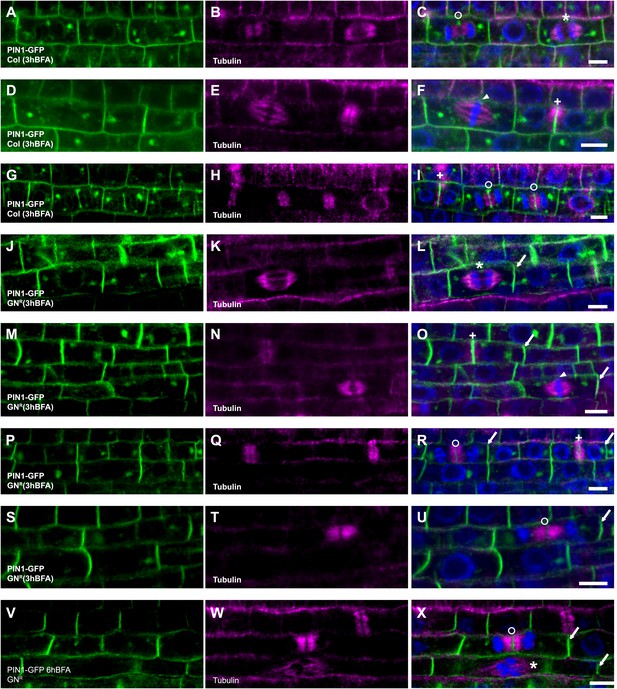
PIN1 recycling in mitotic cells.
BFA-treated seedlings expressing PIN1-GFP (green) were counterstained with tubulin (magenta) and DAPI (blue chromatin). (A–I) In wild-type, PIN1-GFP localizes apolarly at the plasma membrane and at the cell plate in different stages during cytokinesis (BFA 50 μM 3h). (J–U) In BFA-resistant GNOM lines (GNR), PIN1-GFP localizes polarly at the plasma membrane and at the cell plate at the same time (BFA 50 μM 3h). (V–X) Even after prolonged BFA-treatment (6h) of BFA-resistant GNOM seedlings, PIN1 polarity and cell-plate localization are maintained. Mitotic stages: arrowhead, metaphase (panels F and O); asterisk, anaphase (panels C, L, X); circle, telophase (panels C, I, R, U, X); cross, late cytokinesis (panels F, I, O, R). Scale bar, 5 μm.
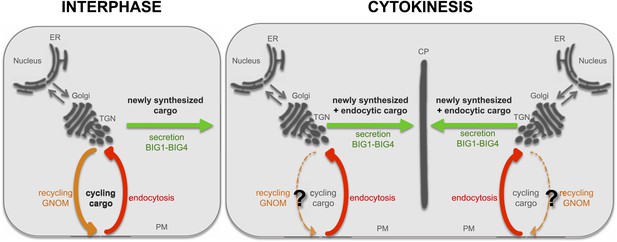
Highly schematic model of secretory and recycling trafficking pathways in interphase and cytokinesis.
In interphase, proteins are synthesized at the ER and are transported via the Golgi to the TGN. The TGN serves as a sorting station. From there, cargo can be transported to the plasma membrane (PM; secretion, green) and this pathway requires the ARF-GEFs BIG1-BIG4. When plasma membrane localized proteins are endocytosed (endocytic pathway, red), they can return to the plasma membrane via the GNOM-dependent recycling pathway (orange). During cytokinesis, newly synthesized proteins are transported from the ER to the Golgi/TGN and from there to the cell plate (CP) in BIG1-BIG4 dependent fashion. Not only newly synthesized cargo but also endocytosed proteins follow this secretory route to the cell plate. PIN1 appears to be exceptional among endocytosed proteins, being recycled to the basal plasma membrane in a GNOM-dependent manner during cytokinesis (?).
Additional files
-
Supplementary file 1
Localization of vesicle trafficking markers.
This table summarizes the localization of different vesicle trafficking markers without BFA (1th column) and with BFA in wild-type (Col; 2th column), big3 (3th column), BFA-resistant GNOM (GNR; 4th column) and BFA-resistant GNOM in big3 mutant background (GNR big3; 5th column). Abbreviations: PM, plasma membrane; CP, cell plate; BFA-comp., BFA-compartment.
- https://doi.org/10.7554/eLife.02131.016





