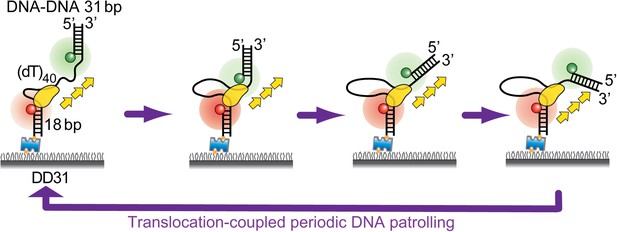
Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA
Figures
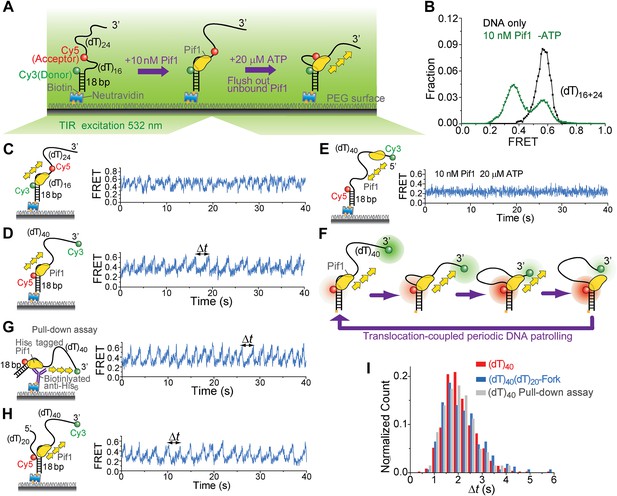
Periodic DNA patrolling by a Pif1 monomer.
(A) A schematic representation of reaction steps for smFRET measurements under the monomer condition. (B) smFRET histograms obtained before and after adding 10 nM Pif1 in the absence of ATP. Peaks centered at EFRET = 0.58 and 0.37 were observed represent 0 and 1 Pif1 molecules binding to DNA, respectively. (C and D), smFRET-time traces showing repetitive translocation of a Pif1 monomer on (dT)40. The labeling positions of Cy3 and Cy5 differ in (C) vs (D). (E) smFRET-time trace for a DNA substrate containing a 5′ Poly(dT) ssDNA overhang (referred to as (dT)40-5′) showing no sawtooth pattern. 10 nM Pif1 and 20 μM ATP were added. (F) A schematic representation of the repetitive looping model. A Pif1 monomer remains bound at the 3′ ss-dsDNA junction and reels in the 3′ ssDNA overhang. (G) A smFRET-time trace showing repetitive translocation of a surface-immobilized Pif1 monomer on non-biotinylated (dT)40. (H) smFRET-time trace showing repetitive looping by a Pif1 monomer on a forked DNA substrate. (I) Histograms of the time interval of repetition (Δt).
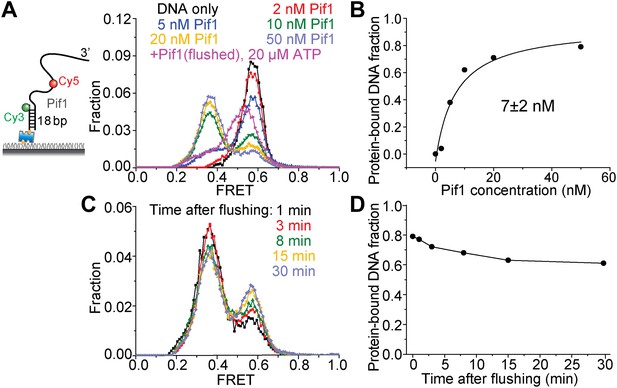
Pif1 binding to (dT)16+24 DNA.
(A) A partial duplex DNA with a 3′ 40-nt Poly(dT) overhang was used to examine Pif1 binding to DNA. Cy3 and Cy5 were attached to the ss–dsDNA junction and the middle of the ssDNA overhang, respectively, separated by 16 nt (this DNA substrate is referred to as (dT)16+24). smFRET histograms were obtained 2 min after introducing the indicated Pir1 concentration to the sample chamber. ATP is absent for all the conditions except for the last condition where the excess Pif1 proteins are flushed. Two distinct peaks centered at EFRET = 0.58 and 0.37 were observed at varying Pif1 concentrations, representing 0 and 1 Pif1 molecules binding to DNA, respectively. (B) Fraction of DNA with Pif1 bound vs Pif1 concentration. (C) Time evolution of the smFRET histogram in the absence of ATP after incubating with 50 nM Pif1 and flushing out the excess unbound proteins (D) Fraction of DNA with Pif1 bound vs time after flushing out the unbound proteins. It should be noted that 7 nM may be larger than the actual KD value, because the 2-min incubation time may not be enough for the system to achieve the equilibrium.

Pif1 binding to (dT)16+24, (dT)40, and (dT)21.
(A) Single molecule time traces for (dT)16+24 DNA only or Pif1-bound (dT)16+24 in the absence of ATP. Unbound proteins were flushed out after incubating 10 nM Pif1 with surface-tethered (dT)16+24. (B) Single molecule time traces for (dT)40 DNA only or Pif1-bound (dT)40 in the absence of ATP. Unbound proteins were flushed out after incubating 10 nM Pif1 with surface-tethered (dT)40. (C) Single molecule time traces of (dT)21 obtained before and after flushing out 10 nMPif1 and at 20 μM ATP. Pif1 binding and dissociation events (marked by black arrows) were observed only if 10 nM Pif1 was maintained in the sample chamber. Sawtooth-shaped patterns appear only when Pif1 binds to (dT)21.
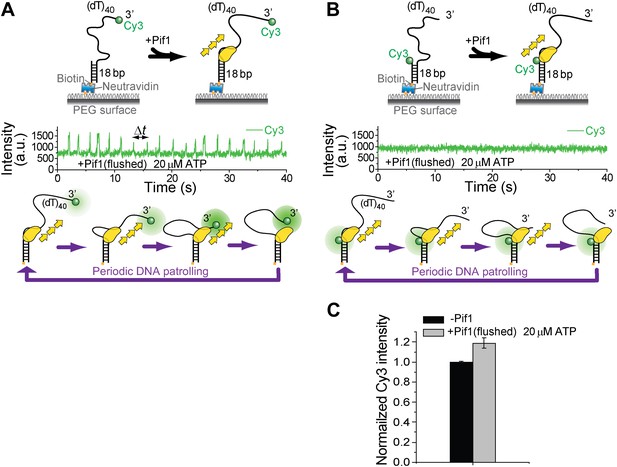
Investigation of Pif1’s periodic patrolling activity using protein-induced fluorescence enhancement (PIFE).
It has been shown that Cy3 intensity would increase as a protein approaches (Hwang et al., 2011). This PIFE effect can be used to infer distance change between Pif1 and the labeled position. (A) When Cy3 is placed at the 3′ end of (dT)40, periodic patterns were observed in the Cy3 intensity time traces under the same monomer condition as in Figure 1D. (B) When Cy3 is placed at the 3′ ss–dsDNA junction of (dT)40, no periodic pattern was observed in the Cy3 intensity time traces under the same monomer condition as in Figure 1D. (C) When Cy3 is placed at the 3′ ss-dsDNA junction of (dT)40, about 20% increase in Cy3 intensity was observed after Pif1 binding to DNA.
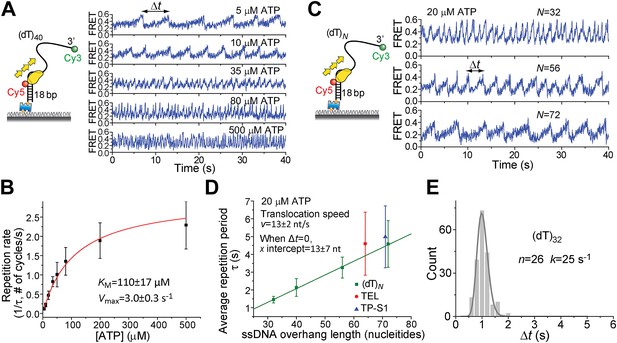
ATP and ssDNA length dependence of the repetition period and step size determination.
(A) smFRET time traces showing Pif1 repetitive looping on (dT)40 at varying ATP concentrations. (B) Michaelis–Menten fit of repetition rate (1/τ) vs ATP concentration. Error bars denote SD. Errors in the fit results are shown in SEM. (C) smFRET time traces of repetitive looping on (dT)N at 20 μM ATP. To obtain the average translocation speed, v, we examined RL with pdDNA containing 3′ Poly(dT) overhangs of varying lengths (N = 32, 40, 56, and 72 nt; referred to as (dT)N). (D) Average repetition period, τ, vs 3′ ssDNA overhang length. The error bar denotes SD. Errors in the fit result are SEM. (E) Δt histogram obtained from a single Pif1 monomer showing 190 repetitive looping events on (dT)32. The solid line is a fit to the Г-distribution.
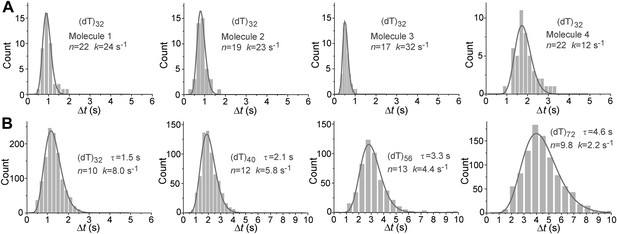
Histograms of the repetition period, Δt, for Pif1 periodic patrolling on (dT)N.
(A) Δt histograms obtained from single Pif1 molecules showing the periodic patrolling behavior on (dT)32. Solid lines are fits with Г-distribution, (Δt)n−1exp(−k·Δt). We found that the translocation speeds (or repetition rates) of single Pif1 translocases differ, and these differences persist during our observation time window of minutes. Similar molecular heterogeneity in the translocation speed was also previously observed for PcrA (Park et al., 2010). (B) Δt histograms obtained from >50 Pif1 molecules showing the periodic patrolling behavior on (dT)N (N = 32, 40, 56, and 72). Due to the molecular heterogeneity, the combined Δt histogram of (dT)32 from an ensemble of single Pif1 molecules is broader than that from a single Pif1 molecule, leading to a smaller n value (∼10) from the Γ-distribution fit. Hence, we used the n values obtained in (A) for the step size determination. Overall, our data consistently place an upper limit on the kinetic step size of <2 nt, and assuming that there exists a defined integer value for the kinetic step size, the kinetic step size of ssDNA translocation is thus 1 nt.
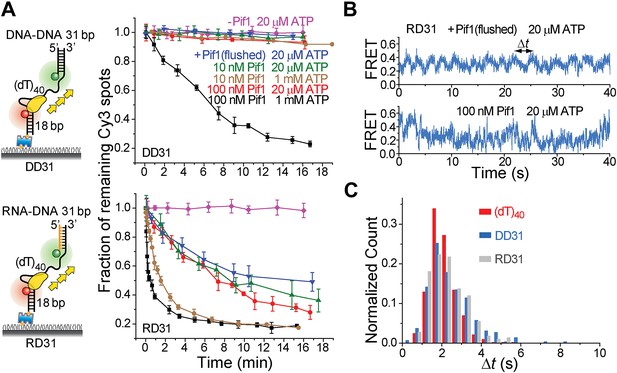
Periodic DNA patrolling of a Pif1 monomer can unwind RNA-DNA hybrids but not dsDNA.
(A) Unwinding curves for DD31 (top) and RD31 (bottom) obtained under different conditions. (B) A smFRET time trace of RD31 obtained under the monomer condition shows Pif1 repetitive looping (top) and a smFRET time trace of RD31 obtained at 100 nM Pif1 shows irregular FRET fluctuations (bottom). (C) Δt histograms for Pif1 periodic patrolling on (dT)40, DD31, and RD31.
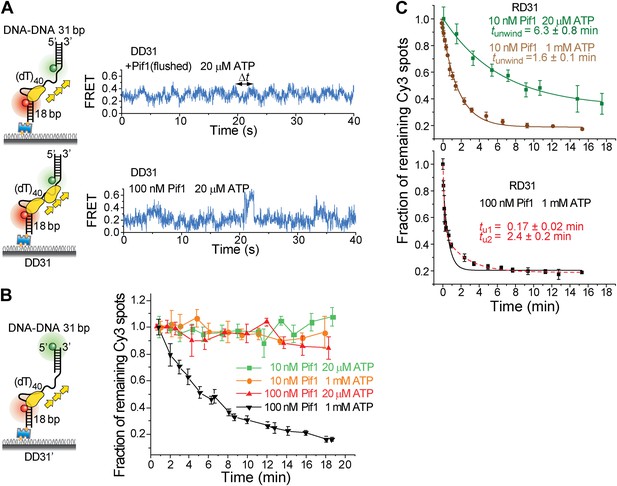
Translocation and unwinding activity of Pif1 on gapped substrates.
(A) Single molecule time traces of Pif1 dynamics on DD31 at two different protein concentrations. After incubating 10 nM Pif1 and flushing out unbound Pif1 (we refer to this condition as the Pif1 monomer condition), single molecule time traces were obtained at 20 μM ATP (top panel). Periodic wave patterns were observed, indicating that a single Pif1 monomer periodically patrols the (dT)40 ssDNA segment. In contrast, in single molecule time traces obtained at 100 nM Pif1 and 20 μM ATP, the regular wave patterns were disrupted (bottom panel), likely due to the binding of multiple Pif1 monomers or protein oligomerization on the substrates at 100 nM Pif1 (Barranco-Medina and Galletto, 2010; Galletto and Tomko, 2013). Similar time traces were obtained under the same conditions for RD31, a gapped substrate containing a 31-bp RNA–DNA hybrid (Figure 3B). (B) Unwinding curves for DD31′ obtained under different conditions. DD31′ is a gapped DNA substrate slightly modified from DD31 by repositioning the Cy3 fluorophore, and is used to test whether the presence of a Cy3 fluorophoe at the ss–dsDNA junction would affect Pif1 unwinding. (C) Unwinding curves for RD31 with exponential fits. Solid lines are single exponential fits. The red dashed line is a bi-exponential fit. At 10 nM Pif1 (the monomer condition), the unwinding curves can be fit well with a single exponential function. However, at 100 nM Pif1, the unwinding curve cannot be fit well with a single exponential function (black solid line), and a bi-exponential function characterized by two unwinding phases (red dashed line) can well describe the curve.
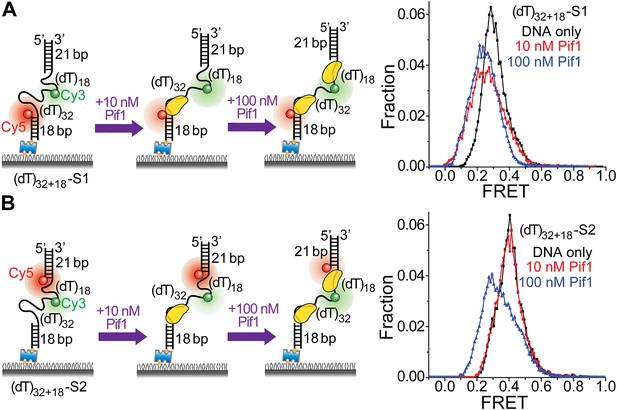
Pif1 monomers are preferentially recruited to 3′ ss-dsDNA junctions.
(A and B) Schematic representations of reaction steps to identify the preferential Pif1 binding site to a gapped DNA substrate. The two gapped DNA substrates used ((dT)32+18-S1 and (dT)32+18-S2) only differ in Cy5 labeling position. smFRET histograms were determined at different Pif1 concentrations.
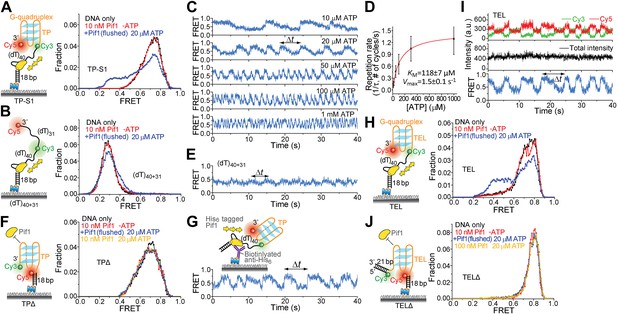
Periodic DNA patrolling of a Pif1 monomer can unwind G4 structures.
(A and B) smFRET histograms of TP-S1 (A) and (dT)40+31 (B) obtained under different conditions. (C) A smFRET time trace showing Pif1 repetitive unwinding on TP-S1 at varying ATP concentrations. (D) Michaelis–Menten fit of the repetition rate (1/τ) vs ATP concentration. Error bars denote SD. Errors in the fit results are SEM. (E) A smFRET time trace showing Pif1 periodic patrolling on (dT)40+31. EFRET changes periodically between ∼0.3 and 0.4, likely due to Pif1 translocation on and off the (dT)31 segment separating Cy3 and Cy5. (F) smFRET histograms for a DNA substrate devoid of the (dT)40 region (referred to as TPΔ) obtained under different conditions. The G4 structure is not unwound when the ssDNA region is absent, likely due to lack of Pif1 loading. (G) A smFRET time trace showing repetitive G4 unwinding of a surface-immobilized Pif1 monomer on non-biotinylated TP-S1. (H and I) smFRET histrograms (H) and time traces (I) of TEL. (J) smFRET histograms of TELΔ obtained under different conditions.
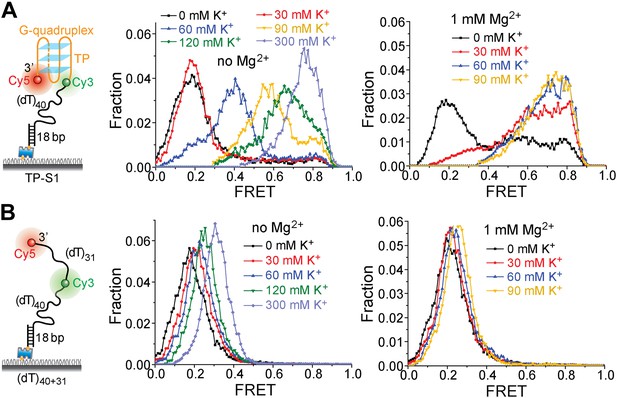
K+ titrations to study G4 formation for the TP sequence in the presence and absence of Mg2+.
(A) smFRET histograms of TP-S1 at varying K+ concentrations in the absence and presence of Mg2+. (B) smFRET histograms of (dT)40+31 at varying K+ concentrations in the absence and presence of Mg2+. TP-S1 and (dT)40+31 only differ in the sequence of the 31-nt ssDNA region flanked by Cy3 and Cy5. Therefore, G4 formation in TP-S1 should result in increased EFRET values compared to the EFRET values of (dT)40+31 at the same [K+]. TP G4 formation is evident when [K+] ≥ 60 mM in the absence of Mg2+, or when [K+] ≥ 30 mM in the presence of Mg2+, indicating that Mg2+ may further stabilize G4 structures in K+. Previous ensemble G4 studies have shown that Mg2+ may facilitate G4 formation (Yan et al., 2010). For (dT)40+31, the FRET peak slightly moves to a higher FRET value, due to increased compaction of ssDNA at higher salt concentrations (Murphy et al., 2004; Chen et al., 2012).
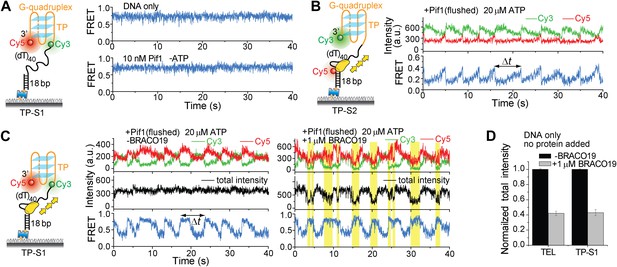
Monomeric Pif1 repetitively unwinds TP G4 in the presence and absence of BRACO19.
(A) smFRET time traces for TP-S1 DNA only and Pif1-bound TP-S1 in the absence of ATP. (B) Single molecule time traces showing periodic DNA patrolling by monomeric Pif1. (C) Single molecule time traces of TP-S1 showing periodic G4 unwinding in the presence and absence of BRACO19. The total fluorescence intensity of Cy3 and Cy5 fluctuates periodically between two levels, and the low intensity level (yellow regions) coincides well with the folded G4 state. The repetitive unwinding is unlikely caused by successive binding of proteins because the excess unbound proteins have been removed from the sample chamber. (D) The average total fluorescence intensity of Cy3 and Cy5 from >10, 000 bare DNA molecules, before and after addition of 1 μM BRACO19. The total fluorescence intensity became 40% of its original level after adding BRACO19 to bare DNA substrates, suggesting that BRACO19 binding to DNA quenches the total fluorescence intensity.
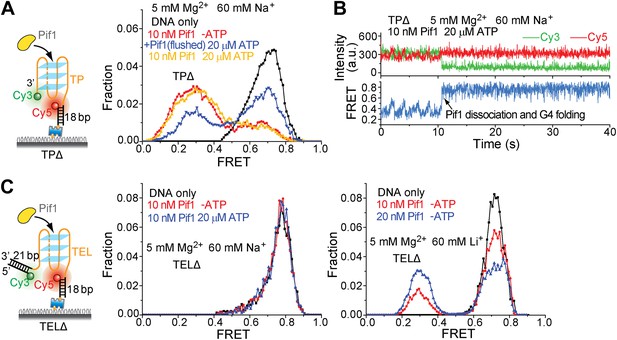
Pif1 binding to TPΔ and TELΔ in the presence of Na+ or Li+.
TPΔ and TEL-DD21 are both partial duplex DNA substrates that contain a G4 sequence but are devoid of the 40-nt ssDNA region at the 5' end of G4 sequence. Figure 5F,J show that Pif1 cannot unwind the G4 structure for both TPΔ and TELΔ in the presence of K+. (A) smFRET histograms of TPΔ obtained under different conditions to study G4 unwinding by Pif1 in Na+. Upon addition of Pif1, similar TP G4 unwinding activity was observed both in the absence and presence of ATP. (B) Single molecule time traces of TPΔ showing an event representing ScPif dissociation and G4 folding obtained after adding 10 nM Pif1 and 20 μM ATP. Sawtooth patterns were observed for TPΔ in the smFRET-time traces, consistent with Pif1's periodic patrolling on unfolded G4 sequence (Figure 2C). The fact that the 0.7 FRET species is more stable in K+ than in Na+ is in line with known G4 stabilizing properties of these monovalent cations and is hence further evidence that the 0.7 FRET peak represents folded G4 molecules. (C) smFRET histograms of TELΔ obtained under different conditions to study G4 unwinding by Pif1 in Na+ or Li+. As in K+, no TEL G4 winding was observed in Na+, but in Li+, TEL G4 winding activity was detected even in the absence of ATP.

K+ and Na+ titrations to study G4 formation for the TEL sequence in the presence and absence of Mg2+.
(A) smFRET histograms of TEL at varying K+ or Na+ concentrations in the absence and presence of Mg2+. (B) smFRET histograms of TEL-DD21 at varying K+ or Na+ concentrations in the presence of Mg2+. The TEL sequence used contains 21 nt, so G4 formation in TEL or TEL-DD21 should result in increased EFRET values compared to the EFRET values of (dT)21DD21 at the same [K+]. TEL G4 formation is nearly complete when [K+] ≥ 30 mM in the absence of Mg2+, and when [K+] ≥ 10 mM in the presence of Mg2+, indicating that Mg2+ may further stabilize G4 in K+. (C) smFRET histograms of (dT)21DD21 at varying K+ concentrations in the presence of Mg2+. The FRET peak of (dT)21DD21 slightly moves to a higher FRET value as [K+] increases, due to increased compaction of ssDNA at higher salt concentrations (Murphy et al., 2004; Chen et al., 2012).
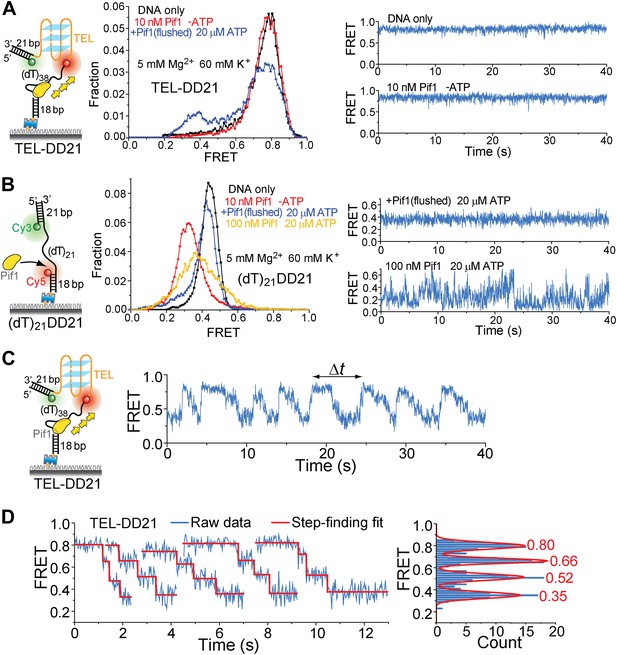
Presence of a DNA duplex next to the 3′ end of the G4 sequence does not affect Pif1′s periodic G4 unwinding activity.
We modified the TEL construct used in Figure 5H by adding a 21-bp DNA duplex next to the 3′ end of the TEL G4 sequence (referred to as TEL-DD21). (A) smFRET histograms of TEL-DD21 obtained under different conditions to study G4 unwinding by Pif1 (left panel). smFRET time traces of TEL-DD21 obtained for DNA only or after adding 10 nM Pif1 in the absence of ATP (right panel). (B) smFRET histograms of (dT)21DD21 obtained under different conditions to study Pif1 binding to (dT)21DD21 (left panel). (dT)21DD21 serves as a control DNA substrate here, because Cy3 and Cy5 were placed at the two ends of a 21-nt Poly(dT) region for (dT)21DD21 and were placed at the two ends of a 21-nt TEL sequence for TEL-DD21. smFRET time traces of (dT)21DD21 were obtained under the monomer condition (flushing out unbound proteins after incubation in 10 nM Pif1) or after adding 100 nM Pif1 and 20 µM ATP (right panel). Under the Pif1 monomer condition (top right), the wave patterns seen for DD31 (Figure 3—figure supplement 1A) were not clearly observed for (dT)21DD21, likely due to a shorter ssDNA region available for Pif's periodic patrolling. However, similar irregular dynamics in the FRET time traces were observed at 100 nM Pif1 and 20 µM ATP as seen for DD31 (bottom right). (C) A representative smFRET time trace showing Pif1 repetitive unwinding on TEL-DD21. Addition of a DNA duplex next to the 3′ end of TEL does not affect the periodic G4 unwinding, indicating that Pif1 monomers unwind G4 but do not go beyond to unwind dsDNA. This is consistent with the findings from the gapped substrates (Figure 3) that the periodic patrolling of a Pif1 monomer cannot unwind dsDNA. (D) The average EFRET values and dwell times at each step (or FRET plateau) were determined by a previously described automated step-finding algorithm (Kerssemakers et al., 2006; Myong et al., 2007; Lee et al., 2012). The distribution of the average EFRET values determined from many unwinding cycles collected from 100–200 molecules is shown in the right panel. The red line shows the Gaussian fit. Peak values are indicated.
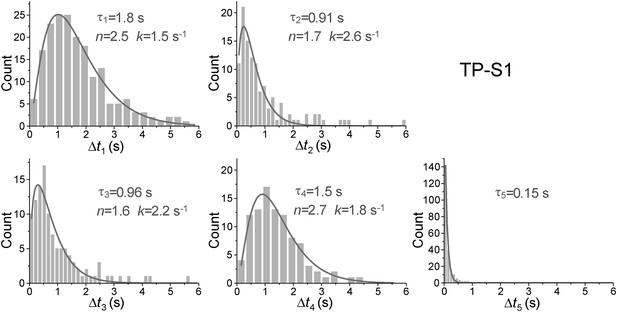
Dwell time histograms of each unwinding step and histogram of G4 refolding time built from TP-S1.
For the Δt1 Δt2, Δt3, and Δt4 histograms (the dwell times for each unwinding step), solid lines are Г-distribution fits. For the Δt5 histogram (the time for the TP G4 structure to refold), the solid line is a single exponential fit.
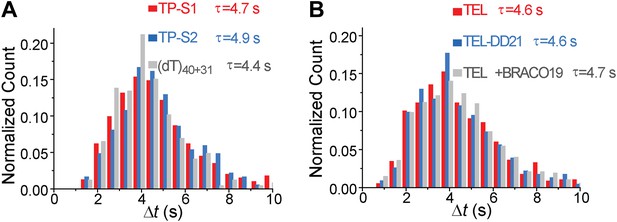
Δt histograms of different DNA substrates.
(A) Histograms of the time interval of repetition (Δt) for TP-S1, TP-S2, and (dT)40+31. (B) Histograms of the time interval of repetition (Δt) for TEL and TEL-DD21. τ is the average repetition period.
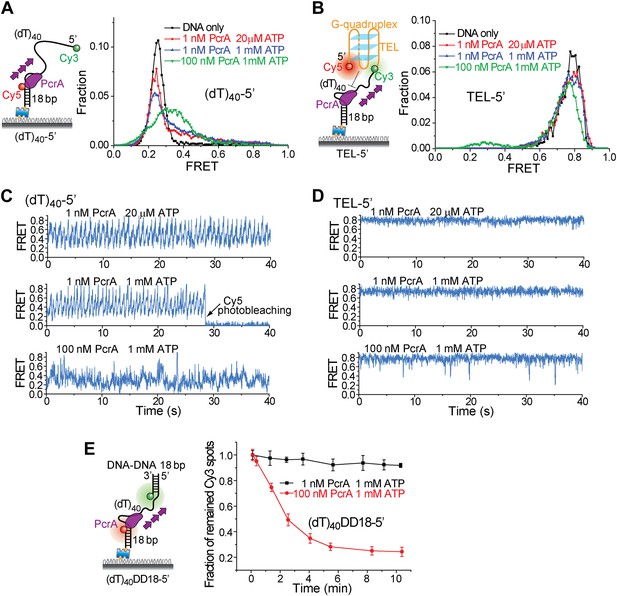
Periodic DNA patrolling of a PcrA monomer cannot unwind G4 structures.
(A) smFRET histograms of (dT)40-5′ obtained for DNA only or at 1 or 100 nM PcrA (ATP was added). (B) smFRET histograms of TEL-5′ obtained for DNA only or at 1 or 100 nM PcrA (ATP was added). (C) Single molecule time traces showing PcrA dynamics on (dT)40-5′ obtained at 1 or 100 nM PcrA. When 1 nM PcrA was added, periodic sawtooth patterns were observed for periodic DNA patrolling by a PcrA monomer as previously demonstrated (Park et al., 2010). However, when 100 nM PcrA was added, the sawtooth patterns were disrupted, and only irregular FRET dynamics were observed, indicating the binding of multiple PcrA monomers per DNA or protein oligomerization. (D) Single molecule time traces of TEL-5′ obtained under the same conditions as in (C). No efficient G4 unwinding was observed. (E) The dsDNA unwinding activity at 1 and 100 nM PcrA. At 1 nM PcrA, no efficient unwinding was observed in 10 min at 1 nM PcrA, whereas efficient unwinding was observed at 100 nM PcrA.
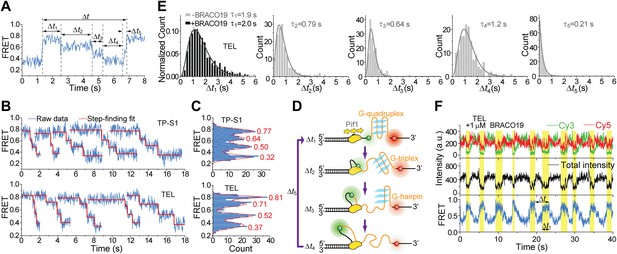
Pif1 unwinds G4 in three discrete steps.
(A) A representative unwinding cycle showing a stepwise pattern for G4 unwinding followed by fast G4 refolding. The dwell times of different parts of the cycle, Δt1, Δt2, Δt3, Δt4 and Δt5, are indicated. (B) The average EFRET values and dwell times at each step were determined by an automated step-finding algorithm (red). (C) Distributions of the average EFRET values determined from many unwinding cycles collected from 100 to 200 molecules. (D) A proposed model for G4 unwinding involving two intermediates. (E) Dwell time histograms of each unwinding step and histogram of G4 refolding time Δt5 (gray). A histogram of Δt1 (F) obtained in the presence of BRACO19 is shown in black. Solid lines are Г-distribution fits. (F) Single molecule time traces showing repetitive G4 winding by a Pif1 monomer in the presence of BRACO19. The total fluorescence intensity of Cy3 and Cy5 fluctuates periodically between two levels, and the low intensity level (yellow regions, Δt1) coincides well with the folded G4 state.
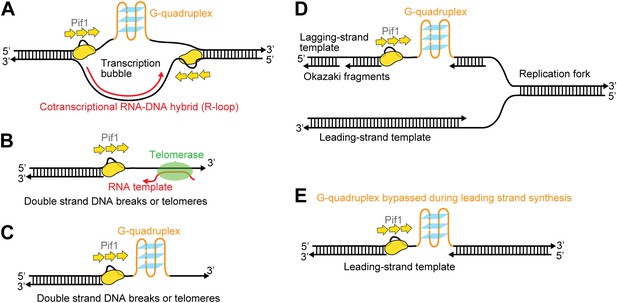
Possible cellular sites for the patrolling activity of a Pif1 monomer.
The periodic patrolling activity may keep Pif1 at its site of in vivo action in resolving biologically relevant ‘R-loops’ (A), displacing telomerase from 3′ ssDNA ends at telomeres or DSBs (B), unwinding G4 structures on the 3′ ssDNA tails at telomeres or DSBs (C), and unwinding G4 structures on the lagging and/or leading strands during DNA replication (D and E).