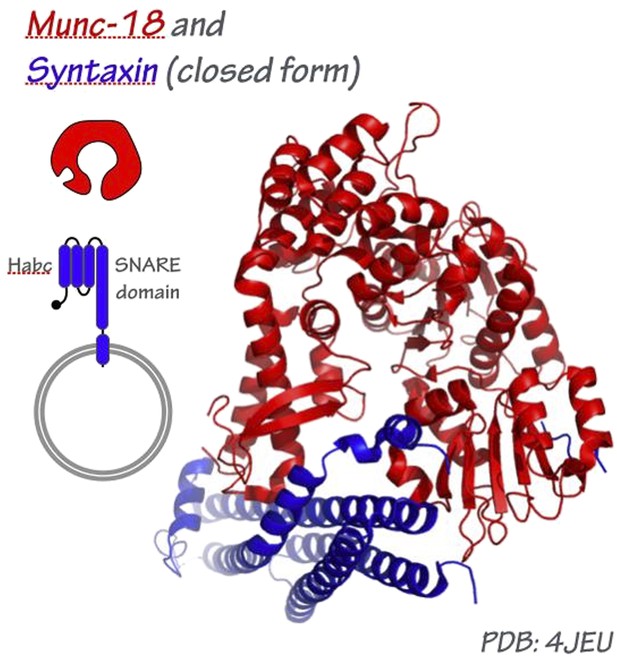
SM proteins Sly1 and Vps33 co-assemble with Sec17 and SNARE complexes to oppose SNARE disassembly by Sec18
Figures
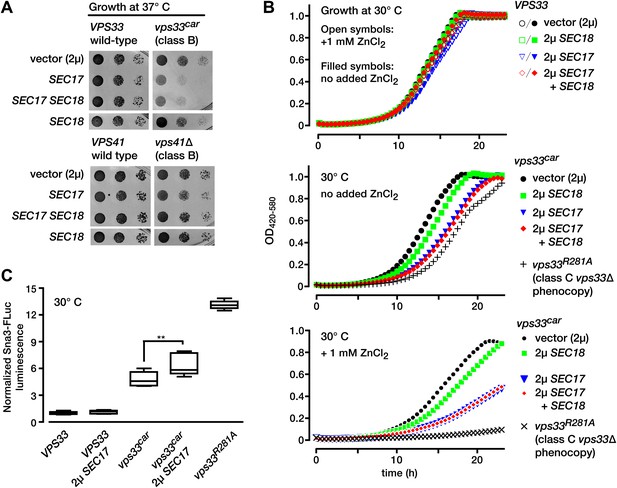
Partial Vps33 deficiency sensitizes cells to overproduction of SNARE disassembly proteins.
(A) Limiting dilution growth assay on synthetic media agar plates incubated at 37°C. (B) Growth curves in selective, synthetic liquid media (YNB lacking uracil and containing 0.05% casamino acids and 2% dextrose, with or without 1 mM ZnCl2). Data points represent the means of n = 4 samples. (C) LUCID analysis of luminal sorting efficiency of the vacuole cargo Sna3-fLuc. Note that vps33car is a hypomorphic allele with partial loss-of-function, while vps33 R281A is a functional null with total loss of function. Box plots summarize n = 5 biological replicates, except for vps33 R281A (n = 4). **p<0.01 (one-way ANOVA). fLuc, firefly luciferase. 2µ, high copy plasmid vector.
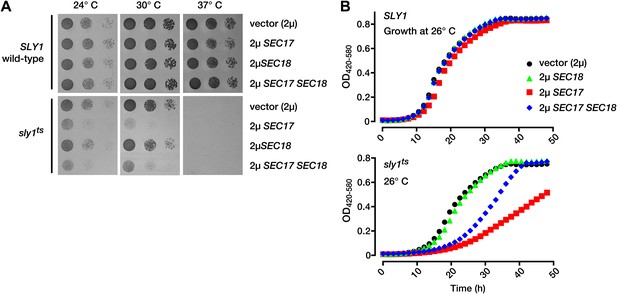
Partial Sly1 deficiency sensitizes cells to overproduction of SNARE disassembly proteins.
(A) Limiting dilution growth assay on plasmid-selective, synthetic media agar plates at 24°, 30° and 37°C. (B) Growth curves of yeast in selective, synthetic liquid media at 26°C. Data points each represent the mean of nine replicate samples. 2µ, high copy plasmid vector.
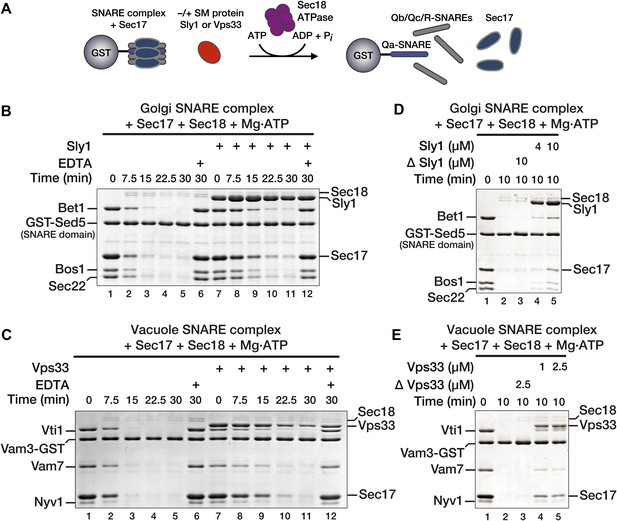
SM proteins oppose Sec18-mediated SNARE disassembly.
(A) Schematic of SNARE disassembly assay. SNARE complexes assembled onto immobilized Qa-SNAREs (Vam3 cytoplasmic domain or Sed5 SNARE domain) were pre-incubated in the presence of Sec17, with or without added SM (Vps33 or Sly1). Sec18 was added to initiate disassembly. The remaining resin-bound material was washed, collected, and analyzed by SDS-PAGE at indicated times. (A and B) Sec17 (20 μM) and Sly1 (10 μM) or Vps33 (2.5 μM) were pre-incubated with SNARE complexes (500 nM) at 30°C for 60 min in Disassembly Buffer. Sec18 (300 nM) was then added. Under these conditions each Sec18 hexamer catalyzed disassembly of >10 SNARE complexes. In negative controls (lanes 6 and 12), Mg2+ was chelated with EDTA prior to Sec18 addition. (D and E) SNARE complex disassembly was assayed as in B and C, but with variable SM protein concentrations as indicated. In ΔSly1 or ΔVps33 lanes, the SM solutions were heated in a boiling water bath for 10 min, plunged into ice-water, and then clarified at 20 k × g to rule out effects of potential heat-stable contaminants in the preparations.
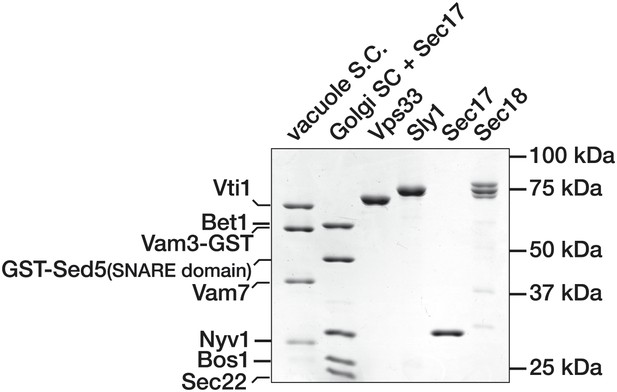
Purified Proteins.
Purified components used in SNARE complex disassembly and binding assays were separated by SDS-PAGE and stained with Coomassie blue.
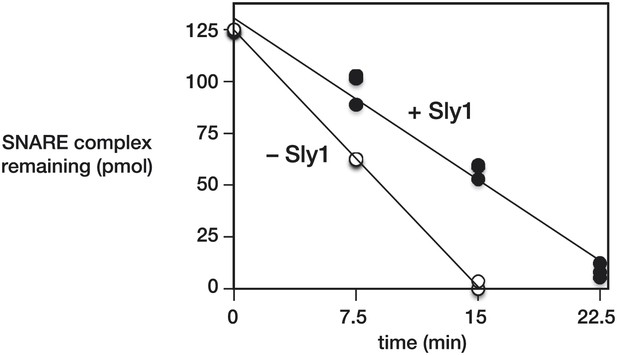
Quantification of SNARE complex protection by Sly1.
Immobilized SNARE complexes (125 pmol in a 250 µl reaction) were pre-incubated for 1 hr at 30°C with saturating Sec17, and with or without Sly1. As in Figure 3, disassembly was initiated by adding Sec18 (12.5 pmol of hexamer). Resin-bound material was washed, collected, and bound proteins were separated by SDS-PAGE and visualized with SYPRO-Ruby. Standard curves of purified proteins were used to determine the amount complex remaining at the indicated times. In the presence of Sly1 the disassembly rate was 0.83 ± 0.04 complexes per Sec18 hexamer per min. In the absence of Sly1 the rate was 1.35 ± 0.01 complexes per Sec18 hexamer per min. Both calculations assume a Sec18 specific activity of 50% and ∼20 turnovers per Sec18 hexamer. Comparable rates of neuronal SNARE disassembly by NSF and α-SNAP (∼1 complex per Sec18 hexamer per min, in 100 mM KCl) were recently reported by Cipriano et al. (2013) (Figure 2G).
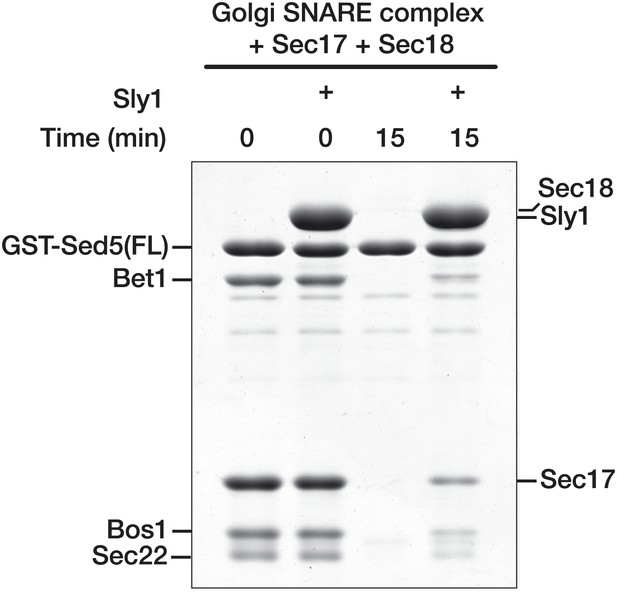
Sly1 protection of SNARE complexes assembled on Sed5 containing Habc domain and N-peptide.
Golgi SNARE complex (500 nM) was assembled on immobilized Sed5 SNARE domain (GST-Sed5FL) containing a Habc domain and N-peptide. SNARE complexes were incubated with Sec17 (20 μM), Sly1 (10 μM), or both for 60 min at 30°C in SM Assay Buffer supplemented with 1 mM ATP and 2 mM MgCl2. 300 nM Sec18 was then added for the indicated period of time, unbound material was washed out, and the bound proteins were analyzed by SDS-PAGE and Coomassie blue staining.
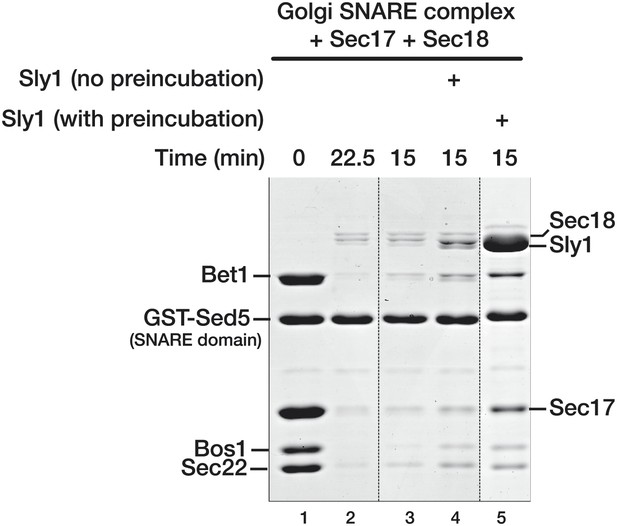
Pre-incubation of Sly1 with SNARE complexes increases the fraction of SNARE complex resistant to Sec18-mediated disassembly.
SNARE complex disassembly was performed as described in Figure 3, with the exception that Sly1 was in the reaction for lane 4 was added simultaneously with Sec18. In lane 5, Sly1 was pre-incubated for the standard 1 hr prior to addition of Sec18. Dashed vertical lines indicate samples from a single experiment, run on parallel gels. Note that GST-Sed5 serves as a loading control.
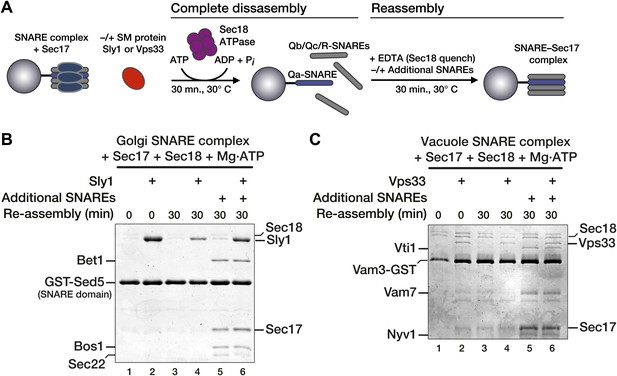
Vps33 and Sly1 do not accelerate SNARE complex re-assembly in solution.
(A) Cartoon schematic of the re-assembly assay. (B and C) SNARE complexes (500 nM) were assembled as in Figure 3. Following pre-incubation of SNARE complexes with 20 μM Sec17 and SM (10 μM Sly1 or 2.5 μM Vps33, as indicated), SNARE complexes were completely disassembled for 30 min by Sec18. Disassembly was terminated with EDTA, and SNARE complex re-assembly was assayed after a further 30 min at 30°C. As indicated, some re-assembly reactions (lanes 5 and 6) were supplemented with soluble SNAREs (Qb, Qc, and R; ∼3 μM each), which were added along with the EDTA quench.
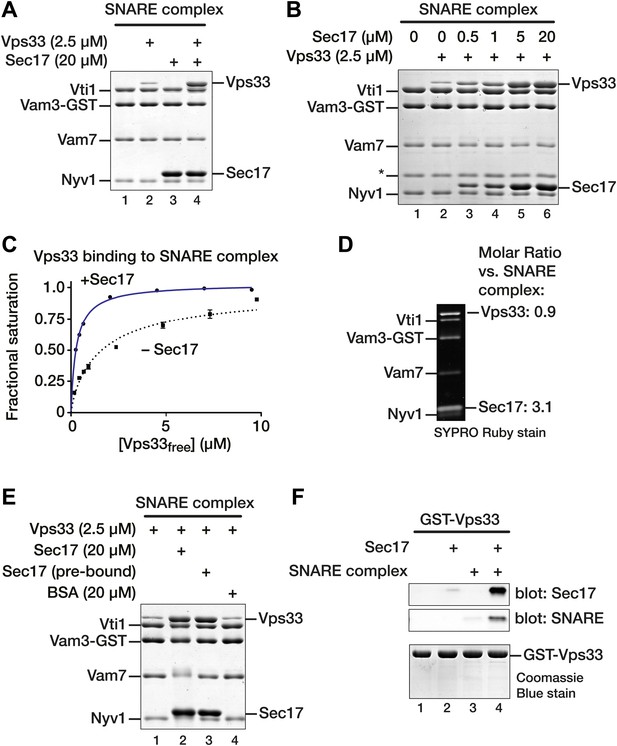
Sec17 promotes Vps33 binding to vacuole SNARE complex.
(A) SNARE complex (500 nM) was assembled on Vam3-GST. Sec17 (20 μM), Vps33 (2.5 μM), or both were incubated with the SNARE complex for 1 hr at 30°C in SM Assay Buffer. Unbound material was washed out, then bound material was separated by SDS-PAGE and visualized with Coomassie blue. (B) The dose–response for Sec17 stimulation of Vps33 binding to SNARE complexes was assayed as in A, but Sec17 concentration was varied (0.5–20 μM) while Vps33 was held constant (2.5 μM). (C) Vps33 binding to SNARE complex with or without Sec17 was assayed as in A and B, except that Vps33 concentration was varied and protein bands were stained and quantified using SYPRO Ruby. The fractional saturation of total Vps33–SNARE complex binding was plotted vs free (total minus bound) Vps33. Fits of a one-site binding model yielded Kdobs = 300 ± 10 nM for Vps33 binding to the SNARE complex in the presence of Sec17, and Kdobs = 1.6 ± 0.10 µM without Sec17. Two-site or cooperative binding models did not substantially improve the fits. (D) To estimate the stoichiometry of SNARE–Sec17–Vps33 binding, complexes were assembled under saturation binding conditions, separated by SDS-PAGE, and analyzed using SYPRO Ruby stain. The band intensities were quantified using standard curves generated with individual purified proteins. (E) Vps33 binding is stimulated by SNARE-associated Sec17. Complexes were assayed in lanes 1 and 2 as in A. In lane 3 (Sec17 pre-bound), Sec17 was bound to SNARE complexes for 60 min at 30°C. Unbound Sec17 was washed out and 2.5 μM Vps33 was then added for an additional 60 min at 30°C. In lane 4, 20 µM BSA was substituted for Sec17. (F) Cooperativity of assembly. GST-Vps33 (500 nM) was immobilized and incubated with 20 μM Sec17, soluble SNARE complex, or both. Bound material was separated by SDS-PAGE and stained with Coomassie blue, or analyzed by immunoblot.
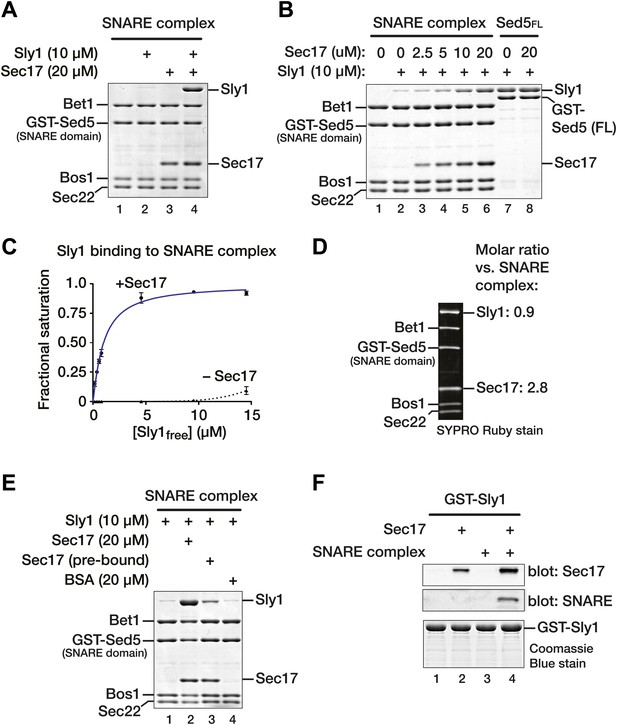
Sec17 promotes Sly1 binding to Golgi SNARE complex.
(A) Golgi SNARE complex (500 nM) was assembled on immobilized Sed5 SNARE domain (GST-Sed5SNARE domain) lacking the N-peptide and Habc segments. SNARE complexes were incubated with Sec17 (20 μM), Sly1 (10 μM), or both for 60 min at 30°C. Unbound proteins were washed out, and bound proteins were separated by SDS-PAGE and stained with Coomassie blue. (B) The dose–response for Sec17 stimulation of Sly1 binding to SNARE complexes was assayed as in A, but Sec17 concentration was varied (2.5–20 μM) while Sly1 was held constant (10 μM). In lanes 7 and 8, the full cytoplasmic domain of Sed5 (Sed5FL, including the Habc and N-peptide segments; 500 nM) was immobilized to test for direct binding of Sec17 to Sed5 and Sly1 in the absence of assembled SNARE complex. (C) Sly1 binding to SNARE complex with or without Sec17 was assayed as in A and B, except that Sly1 concentration was varied and protein bands were stained and quantified using SYPRO Ruby. The fractional saturation of total Sly1-SNARE complex binding was plotted vs free (total minus bound) Sly1. Fits of a one-site binding model yielded with an apparent Kdobs = 1.0 ± 0.1 µM for Sly1 binding to the SNARE complex in the presence of Sec17. It was not possible to fit the no-Sec17 condition. Two-site or cooperative binding models did not substantially improve the fits. (D) Stoichiometry of SNARE-Sec17-Sly1 complexes assembled under saturation conditions was estimated using standard curves of purified proteins of known concentrations. (E) Sly1 binding is stimulated by SNARE-associated Sec17. Binding was assayed in lanes 1 and 2 as in A. In lane 3, Sec17 was pre-bound to SNARE complexes for 60 min at 30°C. Unbound Sec17 was then washed out and Sly1 (10 μM) was added for an additional 60 min at 30°C. In lane, 4 BSA (20 µM) was substituted for Sec17. (F) Cooperativity of assembly. GST-Sly1 (500 nM) was immobilized and incubated with 20 μM Sec17, SNARE complex, or both. Bound material was separated by SDS-PAGE and stained with Coomassie blue or analyzed by immunoblot.
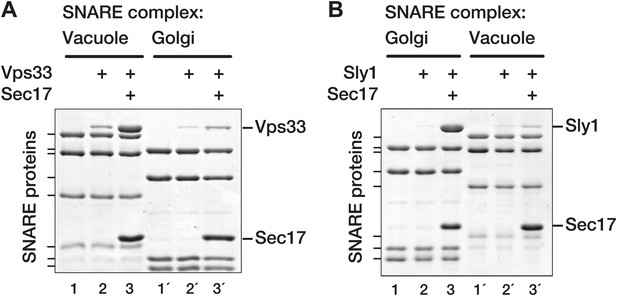
SM proteins touch and recognize cognate SNARE-Sec17 complexes.
Golgi and vacuole SNARE complexes (500 nM) were assembled on affinity supports and assayed for binding in the absence or presence of Sec17 (20 μM). (A) Assay of Vps33 (2.5 μM) binding. (B) Assay of Sly1 (10 µM) binding. Golgi complexes were assembled on Sed5 SNARE domain lacking the N-peptide and Habc segments. Binding reactions were incubated 2 hr at 30°C, unbound material was washed out, and the bound proteins were analyzed by SDS-PAGE and Coomassie blue staining.
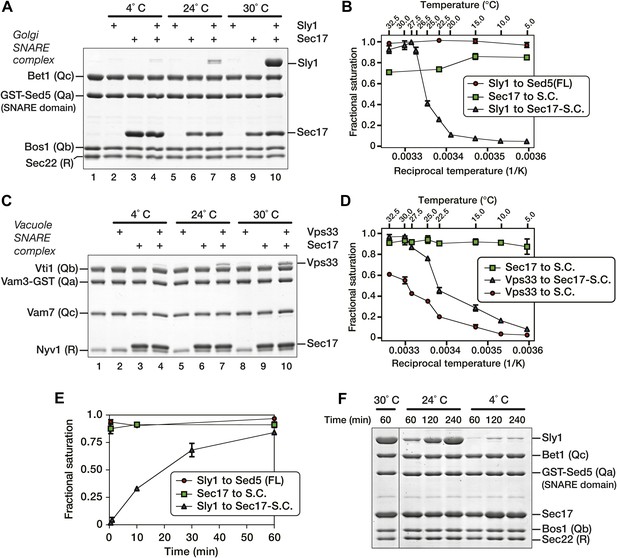
Thermal dependence of SM-SNARE complex association.
(A) Golgi SNARE complexes were assembled on immobilized Sed5 SNARE domain (lacking the Sed5 N-peptide), then assayed for Sly1 (12 µM) binding in the absence and presence of Sec17 (12 µM). The binding reactions were incubated for 60 min at 4°, 24° or 30°C. (B) Sly1 binding to Golgi SNARE complexes (S.C.) was evaluated across a range of temperatures. Note that reciprocal temperature is plotted in units of 1/K, with warmer temperatures on the left side of the plot. Immobilized SNARE complexes (500 nM) were assayed for binding of sub-saturating amounts of Sly1 (8 µM, in the presence of 20 µM Sec17), or for binding of Sec17 alone (7 µM). In an additional control, Sly1 (6 µM) was assayed for binding to the N-peptide of Sed5 (FL; full-length cytoplasmic domain; 500 nM). Bound material was separated by SDS-PAGE, visualized with SYPRO Ruby, and quantified using standard curves of purified proteins of known concentrations. (C) The thermal dependence of Vps33–SNARE association was assayed as in A, except that Vps33 was present at 1 µM and Sec17 was present at 2 µM. (D) Vps33 binding to SNARE complexes was evaluated across a range of temperatures, similar to B. As indicated, Vps33 was present at 1.5 µM and Sec17 was present at 20 µM. (E) The kinetics of Sly1 binding at 30°C to Sec17–SNARE complex were analyzed and quantified using conditions and protein concentrations as in panel B. (F) Sly1 association kinetics are controlled by temperature. Immobilized Golgi SNARE complex (500 nM) was incubated with Sly1 (10 µM) and Sec17 (20 µM) at the indicated temperatures for the indicated times. Bound proteins were separated by SDS-PAGE and stained with Coomassie blue.
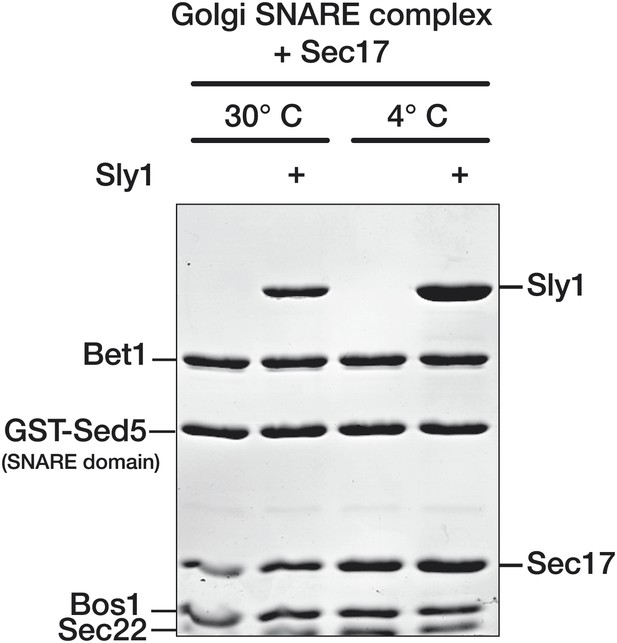
Stability of Sly1-Sec17-SNARE complexes at 4° and 30°C.
Golgi SNARE complex (500 nM) was assembled on immobilized Sed5 SNARE domain (GST-Sed5) lacking the N-peptide and Habc segments. SNARE complexes were incubated with Sec17 (20 μM), Sly1 (10 μM), or both, and incubated for 60 min at 30°C. Unbound protein was washed out and resins were incubated in SM Assay Buffer at either 4° or 30°C for a further 60 min. Resins were washed again at the post-wash incubation temperature (either 4° or 30°C), and the bound proteins were analyzed by SDS-PAGE and Coomassie blue staining.
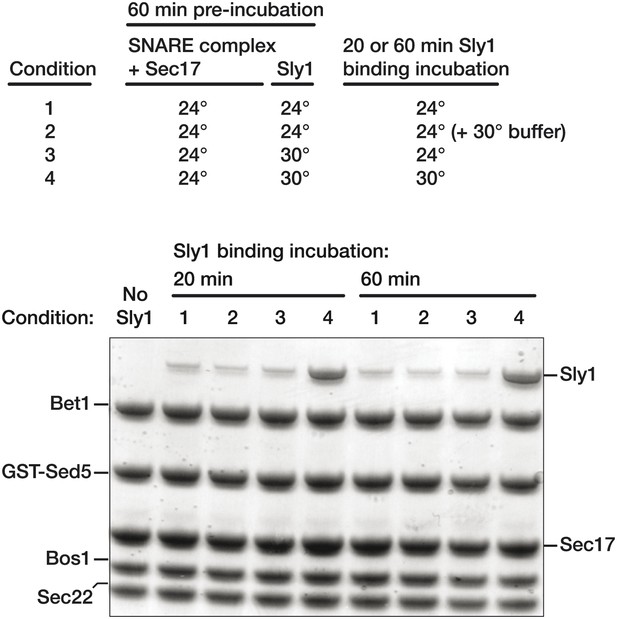
Pre-incubation at 30°C does not trigger conversion of Sly1 into a persistently activated state.
(A) Golgi SNARE complex (500 nM) was assembled on immobilized Sed5 SNARE domain (GST-Sed5) lacking the N-peptide and Habc segments. SNARE complexes were then pre-incubated with Sec17 (20 μM) at room temperature for 60 min prior to addition of Sly1 (10 μM). Sly1 was pre-incubated for 60 min either at room temperature (conditions 1 and 2) or 30 C (conditions 3 and 4). In condition 3, Sly1 pre-incubation for 60 min at 30 C is followed by binding at room temperature (RT). Condition 1 shows baseline binding when Sly1 has only been incubated at RT, while condition 4 shows binding of Sly1 at 30 C (standard experimental condition). To control for the addition of 30°C in condition 4, condition 2 shows binding of Sly1 when the SM has been pre-incubated at RT and an equivalent volume of buffer, added at timepoint zero, was pre-incubated at 30°C.
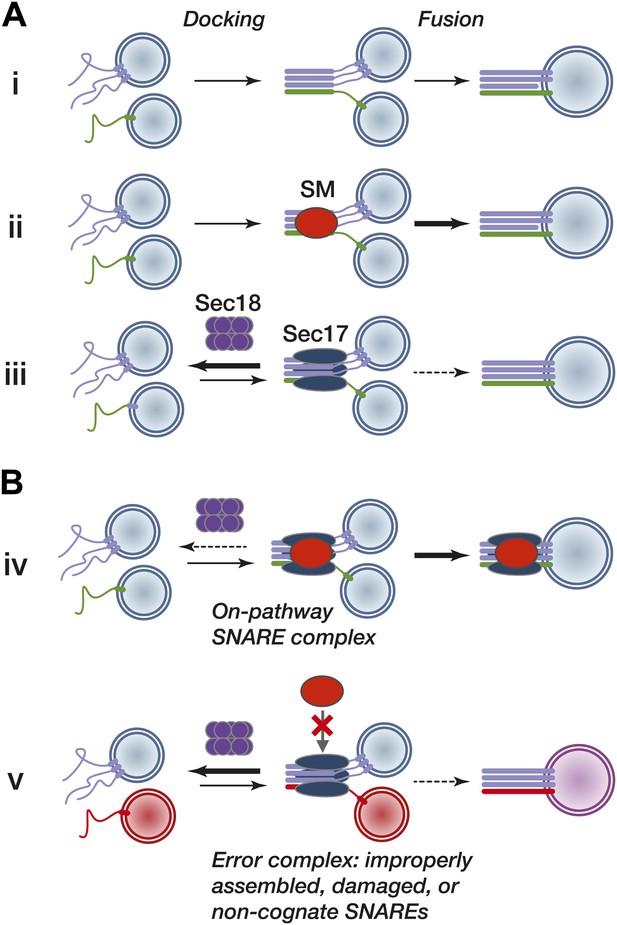
Working model.
(A) Subreactions of SNARE-driven fusion. (i) Basal fusion, as with SNARE proteoliposomes. (ii) SM stimulation of the forward basal fusion reaction. Note that the SM may stimulate trans-complex assembly, the fusogenic activity of extant complexes, or both. (iii) Disassembly of nascent pre-fusion complexes by Sec17 and Sec18 impairs fusion. (B) SM stimulation of fusion in vivo. (iv) Sec17 accelerates SM loading onto cognate SNARE complexes, resulting in more efficient fusion and shielding of the complex from premature disassembly by Sec18. The location of the SM on the SNARE–Sec17 complex, and the dissociation of the SM from the post-fusion complex, are speculative. (v) The SM does not efficiently bind an improperly assembled, damaged, or non-cognate SNARE complex, exposing the complex to kinetic proofreading by Sec17 and Sec18.
Tables
Nomenclature of general and compartment-specific SNAREs and SNARE cofactors employed in this study, and their equivalents in mammalian synaptic exocytosis
| Yeast | Mammal | ||
|---|---|---|---|
| General | General | ||
| AAA-family ATPase | Sec18 | NSF | |
| Sec18 adapter | Sec17 | α-SNAP | |
| Golgi | Vacuole | Synaptic exocytosis | |
|---|---|---|---|
| SM protein | Sly1 | Vps33 | Munc18-1 |
| Qa-SNARE | Sed5 | Vam3 | Syntaxin |
| Qb-SNARE | Bos1 | Vti1 | SNAP-25 (N-domain) |
| Qc-SNARE | Bet1 | Vam7 | SNAP-25 (C-domain) |
| R-SNARE | Sec22 | Nyv1 | Synaptobrevin (VAMP2) |
-
The Q/R taxonomy of SNARE domain subfamilies is derived from Fasshauer et al. (1998).
Yeast lines and plasmids employed in this study
| Name | Genotype | Reference or source |
|---|---|---|
| S. cerevisiae | ||
| SEY6210 | MATα leu2-3112 ura3-52 his3-200 trp1-901 lys2-801 suc2-9 | Robinson et al., 1988 |
| WSY41 | SEY6210; vps41Δ1::LEU2 | Cowles et al., 1997 |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | ATCC |
| BLY3 | BY4742; pep4Δ::KAN VPS33-ttx-GFP::NAT | Lobingier and Merz, 2012 |
| BLY5 | BY4742; pep4Δ::KAN vps33 R281A-ttx-GFP::NAT | Lobingier and Merz, 2012 |
| BLY6 | BY4742; pep4Δ::KAN vps33car [G297V]-ttx-GFP::NAT | Lobingier and Merz, 2012 |
| CBY267 | S288C; MATα ade2-1 ura3-1 trp1-1 leu2-3112 can1-100 | Cao et al., 1998 |
| RSY268 (CBY268) | S288C; MATα ade2-1 ura3-1 trp1-1 leu2-3112 can1-100 sly1ts | Cao et al., 1998 |
| Plasmids | ||
| pDN526 | ApR 2µ URA3 | Nickerson et al., 2012 |
| pDN313 | SEC18 (pDN526) | This study |
| pDN314 | SEC17 (pDN526) | This study |
| pDN315 | SEC17 SEC18 (pDN526) | This study |
| pDN524 | ApR 2µ TRP1 | This study |
| pDN316 | SEC18 (pDN524) | This study |
| pDN317 | SEC17 (pDN524) | This study |
| pDN318 | SEC17 SEC18 (pDN524) | This study |
| pGO735 | ApR CEN LEU2 PGK1pr::RLuc SNA3-FLuc (pRS415) | G Odorizzi (CU-Boulder) |
| pRP1 | pRSF KmR His7-MBP-(tev)- | Lobingier and Merz, 2012 |
| pBL14 | pBL12 KmR VAM3 (1-264)-(tev)-GST | Lobingier and Merz, 2012 |
| pBL19 | pRP1 KmR His7-MBP-(tev)-VTI1 (1-194) | Lobingier and Merz, 2012 |
| pBL20 | pHIS Parallel1 ApR His6-(tev)-NYV1 (1-231) | Lobingier and Merz, 2012 |
| pBL22 | pBL12 KmR His6-GFPA207K-(tev)-Vam7 (190-316) | Lobingier and Merz, 2012 |
| pBL25 | pGST Parallel1 ApR GST-(tev)-SED5SNARE (170-319) | This study |
| pBL26 | pHIS Parallel1 ApR His6-(tev)-BOS1 (1-222) | This study |
| pBL27 | pHIS Parallel1 ApR His6-(tev)-SE22 (1-188) | This study |
| pBL49 | pRP1 KmR His7-MBP-(tev)-Bet1 (1-123) | This study |
| pBL50 | pGST Parallel1 ApR GST-(tev)-SED5SNARE (1-319) | This study |
| pBL51 | pHIS Paralle1 ApR His6-(tev)-Sly1 | This study |
| pSec17 | pTYB12 ApR CBD-(intein)-Sec17 | Schwartz and Merz, 2009 |