Phosphoprotein SAK1 is a regulator of acclimation to singlet oxygen in Chlamydomonas reinhardtii
Figures
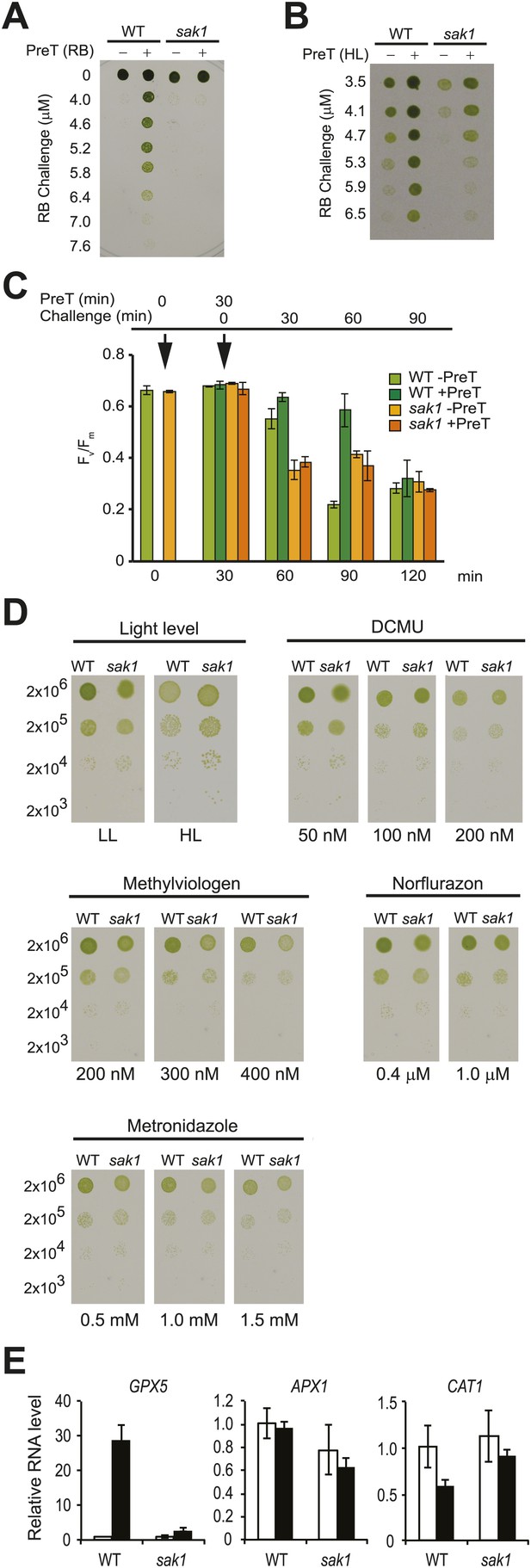
The sak1 mutant is defective in singlet oxygen acclimation.
(A) Acclimation phenotype of WT and sak1. The cells were pretreated in the dark (−) or under light (+) in the presence of rose bengal (RB), which requires light for generation of 1O2. Pretreatment was followed by a subsequent higher concentration of RB (Challenge) as indicated under light. (B) Cells grown in low light were either kept in low light (−) or transferred to high light (+) for an hour before challenge in the light with increasing RB concentrations. (C) Fv/Fm values were measured after each time point indicated. Pretreatment (PreT) with 0.5 μM RB was applied for 30 min with (+PreT) or without (−PreT) light. After the pretreatment, RB was added to both dark and light samples to a final concentration of 3.75 μM RB (challenge), and Fv/Fm was measured for 90 min at 30 min intervals (total 120 min). First arrow: addition of pretreatment; second arrow: addition of challenge. (D) sak1 has wild-type sensitivity to other photo-oxidative stresses. Serial dilutions of WT and sak1 were spotted onto minimal (HS) plates at the indicated light intensity or on TAP plates containing the indicated inhibitor. DCMU, 3-(3,4-dichlorophenyl)-1,1-dimethylurea; low light (LL), 80 µmol photons m−2 s−1; high light (HL), 450 µmol photons m−2 s−1. (E) Gene expression of a known 1O2-responsive gene, GPX5, is induced during acclimation, while two genes associated with H2O2 response, APX1 and CAT1, are not. WT cells were mock-pretreated without RB (white bars) or pretreated with RB in the light (black bars).
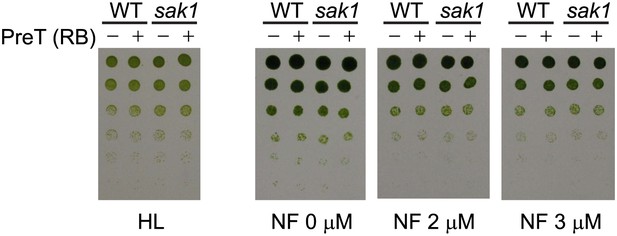
Pretreatment with RB does not increase resistance to high light or norflurazon in cells grown on plates.
Cells were pretreated with 1 μM RB with (+) or without (−) light, then spotted on minimal plates and grown under high light (HL) or grown photoheterotrophically on TAP plates containing norflurazon (NF) and grown under low light for 4 days. Cells were spotted in serial dilutions.
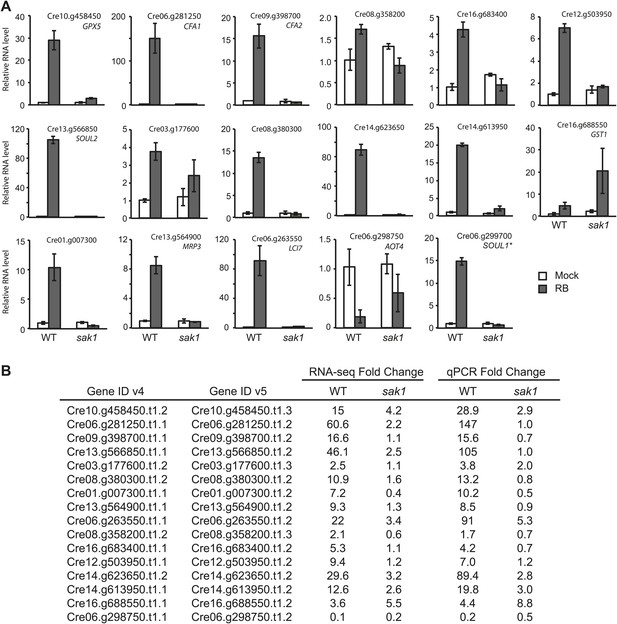
qRT-PCR analysis of genes identified to be 1O2-responsive by RNA-seq.
(A) The error bars indicate standard deviation of biological triplicates. The locus of the transcript (v5) and gene name if annotated, are indicated. *SOUL1 was named gene in v4 but not in v5. (B) Comparison of fold change values from RNA-seq data and qPCR. Fold change values were calculated for RNA-seq as described in ‘Material and methods’, and the values for qPCR are averages obtained from biological triplicates.
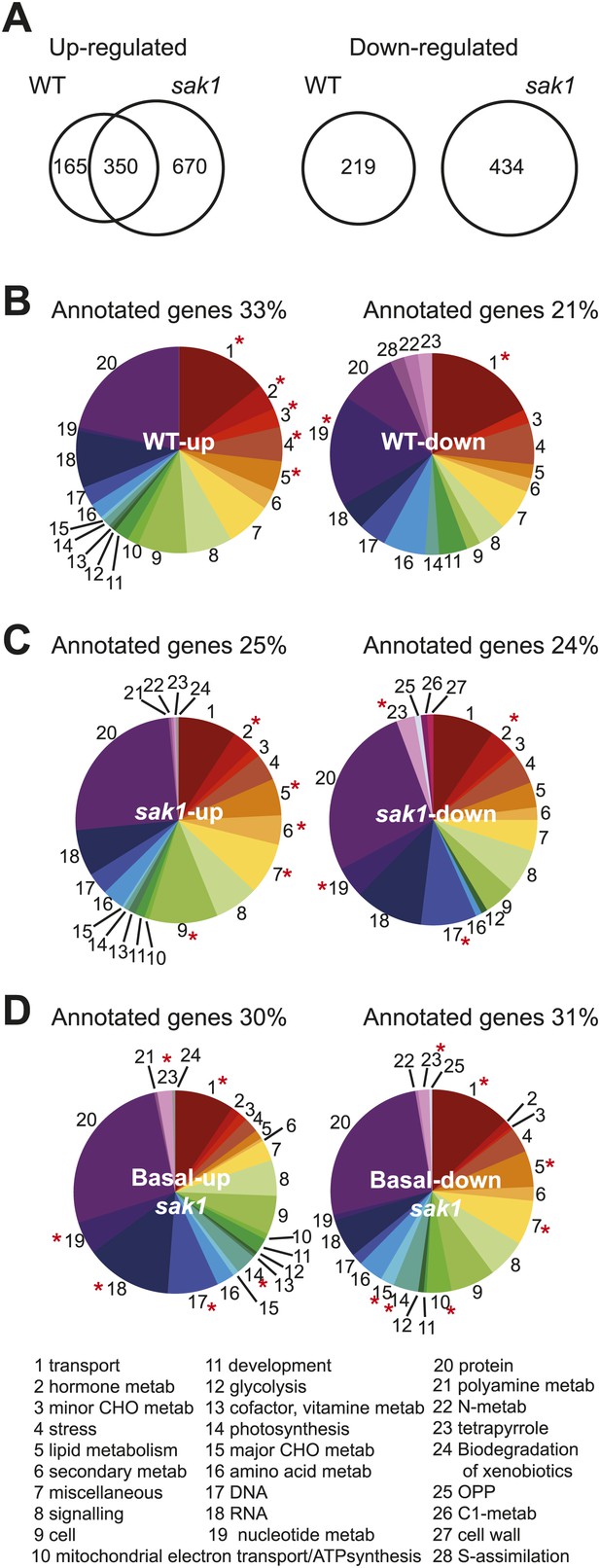
Differentially expressed genes from pair-wise comparisons.
(A) Venn diagram representing differentially expressed genes in WT and sak1. Mapman functional classes distribution of differentially expressed genes (passing criteria of fold change greater than 21 [up] or smaller than 2−1 [down] with FDR <1%) during acclimation in (B) WT and (C) sak1. (D) Differentially expressed genes when comparing WT and sak1 in basal conditions (i.e., before exposure to 1O2). The functional classes represented by the numbers are listed; asterisks indicate classes that were enriched compared to the genome.
Genetic and molecular analysis of sak1.
(A) The insertion of a zeocin resistance gene and the RB sensitivity phenotype are linked. Twelve complete tetrads from a backcross of sak1 to wild type are shown. Numbers indicate independent tetrads, and letters (a-d) indicate the individual progeny from tetrads. (B) Gene structure of SAK1 and the insertion site. Gray boxes indicate positions of primers used for qPCR. (C) Transformation of sak1 with a genomic fragment containing SAK1 rescues the acclimation phenotype. sak1(gSAK1)-1 and sak1(gSAK1)-2 are two independent transformants. (D) sak1(gSAK1)-1 and sak1(gSAK1)-2 show recovery of 1O2 target gene expression. Y-axis indicates fold change during acclimation to 1O2. (E) qRT-PCR of SAK1 in WT and sak1 mutant using primers for 5′- and 3′-UTR shown in panel B. (F) SAK1 protein is induced in WT and detected as higher molecular weight bands during acclimation to 1O2 generated by RB. (G) SAK1 transcript probed for 5′-UTR in cells transferred from low light to high light for 1 hr. Error bars indicate standard deviation of biological triplicates.
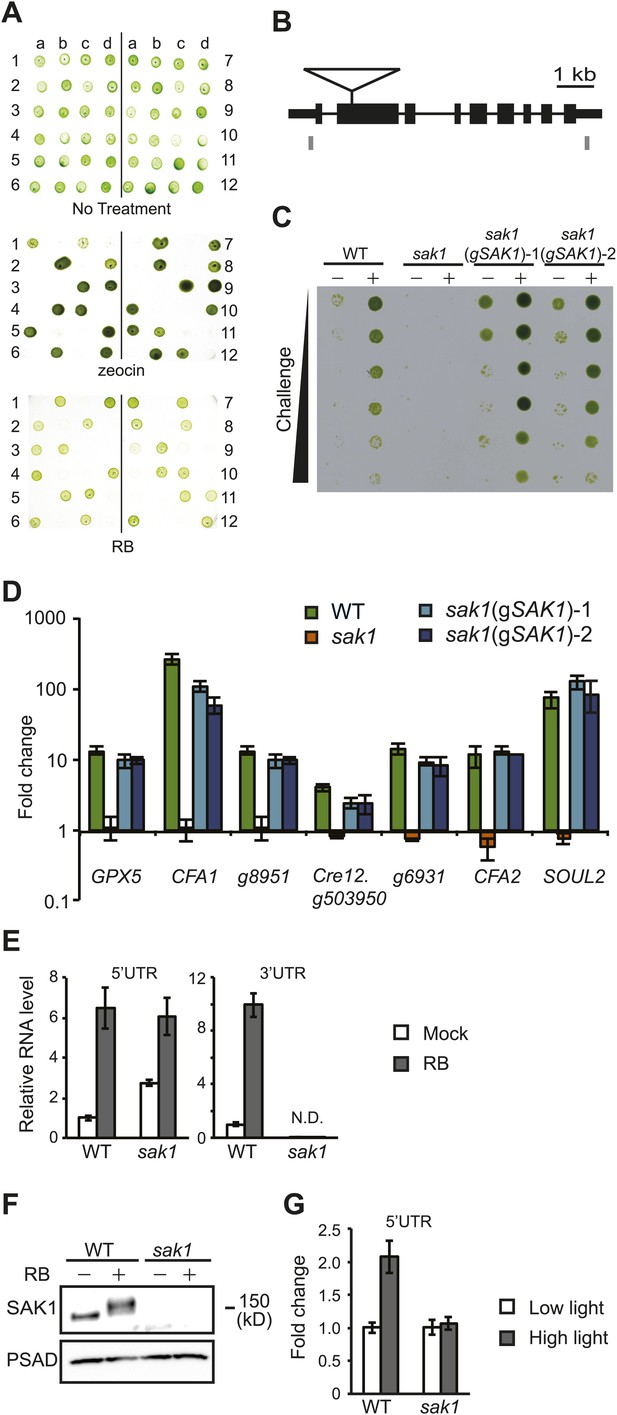
SAK1 contains an uncharacterized domain present in some bZIP transcription factors.
Schematic of relative positions of SAK1 and bZIP domains. One protein (Cv28) contains a mitochondrial termination factor (mTERF) domain. The letters and numbers in the abbreviated names represent initials of the species and numbers listed in Table 8. Proteins with italicized names contain bZIP domains that were recognized by Pfam but scored below significance.
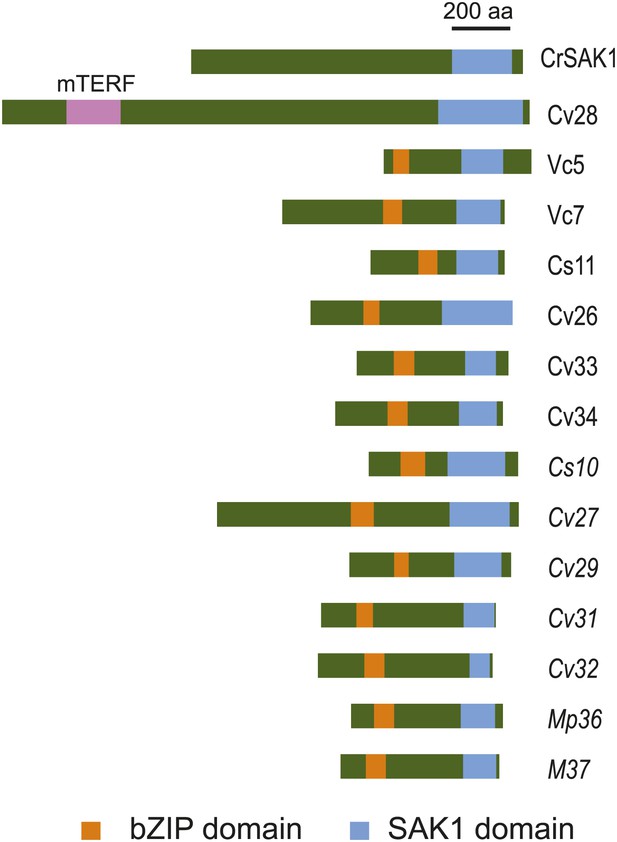
Multiple sequence alignment of SAK1 domains.
The SAK1 domains of 37 chlorophyte proteins were aligned by MUSCLE (phylogeny.fr). Protein identities are as shown in Table 8. Star indicates a relatively conserved residue within the SAK1 domain that was predicted to be a possible phosphorylation site (Figure 5—figure supplement 3).
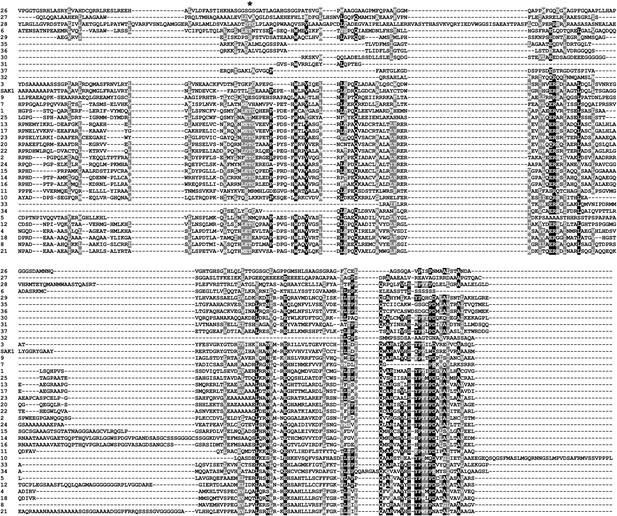
Secondary structure prediction of SAK1 domain.
SAK1 domain modeled against its best-hit nickel cobalt resistance protein cnrr by PHYRE. 44% (coverage) of the SAK1 domain was aligned with 73.6% confidence.
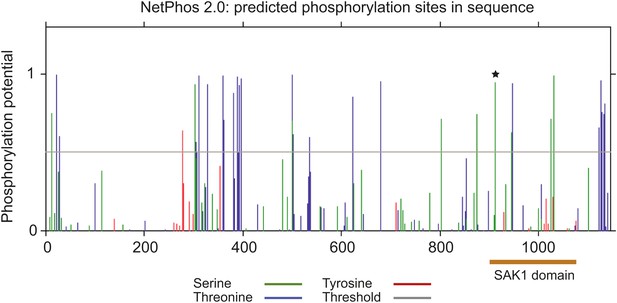
Prediction of phosphorylation sites in SAK1.
Prediction of phosphorylation sites by NetPhos 2.0. Orange bar indicates the position of SAK1 domain, star indicates a relatively conserved residue among the 37 members containing the SAK1 domain.
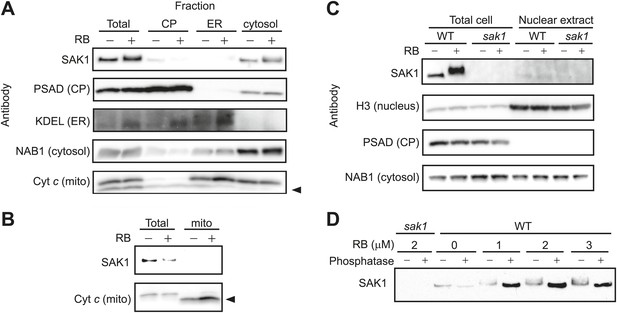
SAK1 is a phosphorylated protein that is in the cytosol.
(A and B) SAK1 is detected in the cytosol and not in other subcellular fractions. (C) SAK1 is not enriched in nuclear extracts. Approximately 30 μg of protein was loaded into each well except for mitochondrial fractions that were loaded approximately 7.5 μg protein due to low protein yield in isolated fractions. Subcellular markers: Chloroplast (CP), PSAD; Endoplasmic reticulum (ER), KDEL; Cytosol, NAB1; Mitochondria (mito), cytochrome c (Cyt c); Nuclear, histone 3 (H3). The arrowhead indicates the band corresponding to Cyt c. (D) Protein extracts from cells treated with increasing concentrations of RB were then treated with phosphatase (+) or only with buffer (−) before detection of SAK1 by immunoblot analysis.
Tables
Moderate induction of 1O2 genes during high light exposure
| Fold change (SD)* | ||
|---|---|---|
| Gene name or ID | WT | sak1 |
| GPX5 | 2.86 (1.06) | 1.08 (0.23) |
| CFA1 | 3.75 (0.99) | 1.78 (0.52) |
| SOUL2 | 3.45 (1.25) | 1.82 (0.22) |
| MRP3 | 3.10 (0.39) | 2.37 (0.32) |
| Cre14.g613950 | 1.42 (0.53) | 1.57 (0.46) |
| LHCSR1† | 14.91 (4.25) | 2.91 (1.35) |
-
*
Fold change values are the average of biological triplicates and their standard deviations are indicated in parentheses.
-
†
Known to have elevated expression in high light grown cells (Peers et al., 2009).
Expression of H2O2 response genes during 1O2 acclimation
| Gene ID | RPKM* | Fold change† | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene name | v4 | v5 | WT-mock | WT-RB | sak1-mock | sak1-RB | WT | sak1 |
| APX1 | Cre02.g087700.t1.1 | Cre02.g087700.t1.2 | 49.70 | 36.22 | 79.65 | 58.83 | 0.73 | 0.74 |
| MSD3 | Cre16.g676150.t1.1 | Cre16.g676150.t1.2 | 0.30 | 0.18 | 0.70 | 0.17 | 0.60 | 0.25 |
| MDAR1 | Cre17.g712100.t1.1 | Cre17.g712100.t1.2 | 35.95 | 38.30 | 33.53 | 51.34 | 1.07 | 1.53 |
| DHAR1 | Cre10.g456750.t1.1 | Cre10.g456750.t1.2 | 20.40 | 40.93 | 25.69 | 42.18 | 2.01 | 1.64 |
| GSH1 | Cre02.g077100.t1.1 | Cre02.g077100.t1.2 | 28.27 | 26.91 | 40.42 | 49.95 | 0.95 | 1.24 |
| GSHR1 | Cre06.g262100.t1.2 | Cre06.g262100.t1.3 | 19.17 | 19.02 | 19.39 | 22.41 | 0.99 | 1.16 |
| VTC2 | Cre13.g588150.t1.1 | Cre13.g588150.t1.2 | 18.16 | 62.53 | 35.10 | 103.12 | 3.44 | 2.94 |
-
*
Average of RPKM obtained from two sequencing lanes as described in ‘Material and methods’.
-
†
Calculated as ratio of (RPKM-RB) / (RPKM-mock).
Enriched functional classes among differentially expressed genes in WT during 1O2 acclimation
| Primary MapMan class | Secondary Mapman class | Gene ID (v4) | Gene ID (v5) | Gene name | Annotation |
|---|---|---|---|---|---|
| Up-regulated genes | |||||
| transport | ABC transporters and multidrug resistance systems | Cre03.g169300.t1.1 | Cre03.g169300.t2.1 | ABC transporter (ABC-2 type) | |
| Cre04.g220850.t1.1 | Cre04.g220850.t1.2 | ABC transporter (ABC-2 type) | |||
| Cre11.g474600.t1.1§ | Cre02.g095151.t1 | ABC transporter (ABC-2 type) | |||
| Cre03.g151400.t1.2 | Cre03.g151400.t1.3 | ABC transporter (subfamilyA member3) | |||
| Cre14.g618400.t1.1§ | Cre14.g618400.t1.2 | ABC transporter | |||
| Cre09.g395750.t1.2 | Cre09.g395750.t1.3 | ABC transporter (plant PDR pleitropic drug resistance) | |||
| Cre14.g613950.t1.1§ | Cre14.g613950.t2.1 | ABC transporter, Lipid exporter ABCA1 and related proteins | |||
| Cre17.g725150.t1.1 | Cre17.g725150.t1.2 | ABC transporter | |||
| Cre04.g224400.t1.2§ | Cre04.g224400.t1.3 | ABC transporter (plant PDR pleitropic drug resistance) | |||
| Cre13.g564900.t1.1§ | Cre13.g564900.t1.2 | MRP3 | ABC transporter, Multidrug resistance associated protein | ||
| Cre17.g721000.t1.1 | Cre17.g721000.t1.2 | ABC transporter (ABCA) | |||
| Cre04.g224500.t1.2 | Cre04.g224500.t1.3 | ABC transporter (plant PDR pleitropic drug resistance) | |||
| Cre01.g007000.t1.1§ | Cre01.g007000.t1.2 | ABC transporter (ABC-2 type) | |||
| unspecified anions | Cre13.g574000.t1.2 | Cre13.g574000.t1.3 | Chloride channel 7 | ||
| Cre17.g729450.t1.1 | Cre17.g729450.t1.2 | Chloride channel 7 | |||
| amino acids | Cre04.g226150.t1.2 | Cre04.g226150.t1.3 | AOC1 | Amino acid carrier 1; belongs to APC (amino acid polyamine organocation) family | |
| misc | Cre16.g683400.t1.1§ | Cre16.g683400.t1.2 | CRAL/TRIO domain (Retinaldehyde binding protein-related) | ||
| Cre17.g718100.t1.1 | Cre17.g718100.t1.2 | Phosphatidylinositol transfer protein SEC14 and related proteins (CRAL/TRIO) | |||
| Cre06.g311000.t1.2 | Cre06.g311000.t1.3 | FBT2 | Folate transporte | ||
| calcium | Cre09.g410050.t1.1§ | Cre09.g410050.t1.2 | Ca2+ transporting ATPase | ||
| potassium | Cre07.g329882.t1.2 | Cre07.g329882.t1.3 | Ca2+-activated K+ channel proteins | ||
| phosphate | Cre16.g686750.t1.1 | Cre16.g686750.t1.2 | PTA3 | Proton/phosphate symporter | |
| metal | Cre13.g570600.t1.1 | Cre13.g570600.t1.2 | CTR1 | CTR type copper ion transporter | |
| metabolite transporters at the mitochondrial membrane | Cre06.g267800.t1.2 | Cre06.g267800.t2.1 | Mitochondrial carrier protein | ||
| hormone metabolism* | brassinosteroid | Cre16.g663950.t1.1 | Cre16.g663950.t1.2 | Sterol C5-desaturase | |
| Cre02.g076800.t1.1 | Cre02.g076800.t1.2 | delta14-sterol reductase | |||
| Cre12.g557900.t1.1 | Cre12.g557900.t1.1 | CDI1 | C-8,7 sterol isomerase | ||
| Cre02.g092350.t1.1 | Cre02.g092350.t1.2 | Cytochrome P450, CYP51 Sterol-demethylase | |||
| Cre12.g500500.t1.2 | Cre12.g500500.t2.1 | SAM-dependent methyltransferases | |||
| jasmonate | Cre19.g756100.t1.1 | Cre03.g210513.t1 | 12-oxophytodienoic acid reductase | ||
| auxin | Cre14.g609900.t1.1 | Cre14.g609900.t1.1 | Predicted membrane protein, contains DoH and Cytochrome b-561/ferric reductase transmembrane domains | ||
| Cre06.g276050.t1.1 | Cre06.g276050.t1.2 | Aldo/keto reductase | |||
| Cre16.g692800.t1.2 | Cre16.g692800.t1.3 | Aldo/keto reductase | |||
| Cre03.g185850.t1.2 | Cre03.g185850.t1.2 | pfkB family, sugar kinase-related | |||
| minor CHO metabolism | others | Cre06.g276050.t1.1 | Cre06.g276050.t1.2 | Aldo/keto reductase | |
| Cre16.g692800.t1.2 | Cre16.g692800.t1.3 | Aldo/keto reductase | |||
| Cre03.g185850.t1.2 | Cre03.g185850.t1.2 | pfkB family, sugar kinase-related | |||
| callose | Cre06.g302050.t1.1 | Cre06.g302050.t1.2 | 1,3-beta-glucan synthase | ||
| myo-inositol | Cre03.g180250.t1.1 | Cre03.g180250.t1.2 | Myo-inositol-1-phosphate synthase | ||
| stress | biotic | Cre01.g057050.t1.1§ | Cre03.g144324.t1 | Leucine Rich Repeat | |
| Cre01.g016200.t1.2 | Cre01.g016200.t1 | Mlo Family | |||
| Cre28.g776450.t1.1§ | Cre08.g358573.t1 | PSMD10 | 26S proteasome regulatory complex | ||
| abiotic | Cre12.g501500.t1.1 | NF† | |||
| Cre02.g132300.t1.2 | Cre09.g395732.t1 | DnaJ domain | |||
| Cre07.g339650.t1.2 | Cre07.g339650.t1.3 | DNJ20 | DnaJ-like protein | ||
| Cre01.g033300.t1.1§ | Cre01.g033300.t2.1 | No annotation‡ | |||
| Cre16.g677000.t1.1 | Cre16.g677000.t1.2 | HSP70E | Heat shock protein 70E | ||
| Cre08.g372100.t1.1 | Cre08.g372100.t1.2 | HSP70A | Heat shock protein 70A | ||
| lipid metabolism | phospholipid synthesis | Cre13.g604700.t1.2 | Cre13.g604700.t1.3 | PCT1 | CDP-alcohol phosphatidyltransferase/Phosphatidylglycerol-phosphate synthase |
| Cre06.g281250.t1.1§ | Cre06.g281250.t1.2 | CFA1 | Cyclopropane fatty acid synthase | ||
| Cre09.g398700.t1.1§ | Cre09.g398700.t1.2 | CFA2 | Cyclopropane fatty acid synthase | ||
| ‘exoticsߣ (steroids, squalene etc) | Cre01.g061750.t1.1 | Cre03.g146507.t1 | SPT2 | Serine palmitoyltransferase | |
| Cre83.g796250.t1.1 | NF† | SPT1 | Serine palmitoyltransferase | ||
| Cre02.g137850.t1.1 | Cre09.g400516.t1 | TRAM (translocating chain-associating membrane) superfamily | |||
| FA synthesis and FA elongation | Cre03.g182050.t1.1 | Cre03.g182050.t1 | Long-chain acyl-CoA synthetases (AMP-forming) | ||
| Cre06.g256750.t1.1 | Cre06.g256750.t1.2 | Acyl-ACP thioesterase | |||
| misc | short chain dehydrogenase/reductase (SDR) | Cre12.g556750.t1.2 | Cre12.g556750.t1.3 | Short chain dehydrogenase | |
| Cre27.g775000.t1.1 | Cre12.g549852.t1 | Short chain dehydrogenase | |||
| Cre17.g731350.t1.2 | Cre17.g731350.t1.2 | Short chain dehydrogenase | |||
| Cre08.g381510.t1.1§ | NF† | Short chain alcohol dehydrogenase | |||
| UDP glucosyl and glucoronyl transferases | Cre02.g144050.t1.1 | Cre02.g144050.t2.1 | Acetylglucosaminyltransferase EXT1/exostosin 1 | ||
| Cre16.g659450.t1.1 | Cre16.g659450.t1.2 | Lactosylceramide 4-alpha-Galactosyltransferase | |||
| Cre03.g173300.t1.1 | Cre03.g173300.t1.2 | Lactosylceramide 4-alpha-Galactosyltransferase | |||
| dynamin | Cre02.g079550.t1.1 | Cre02.g079550.t1.2 | Dynamin-related GTPase, involved in circadian rhythms | ||
| misc2 | Cre06.g258600.t1.1§ | Cre06.g258600.t2.1 | Predicted hydrolase related to dienelactone hydrolase | ||
| acid and other phosphatases | Cre06.g249800.t1.1 | Cre06.g249800.t1.2 | Sphingomyelin synthase | ||
| Down-regulated genes | |||||
| nucleotide metabolism | salvage | Cre13.g573800.t1.1 | Cre13.g573800.t1.2 | Phosphoribulokinase / Uridine kinase family | |
| synthesis | Cre12.g503300.t1.1 | Cre12.g503300.t1.2 | Phosphoribosylamidoimidazole-succinocarboxamide synthase | ||
| Cre06.g308500.t1.1 | Cre06.g308500.t1.2 | CMP2 | Carbamoyl phosphate synthase, small subunit | ||
| Cre14.g614300.t1.1 | Cre14.g614300.t1.2 | Inosine-5-monophosphate dehydrogenase | |||
| transport | ABC transporters and multidrug resistance systems | Cre06.g273750.t1.2 | Cre06.g273750.t1.3 | SUA1 | Chloroplast sulfate transporter |
| Cre02.g083354.t1.1 | Cre02.g083354.t1 | ATP-binding cassette, subfamily B (MDR/TAP), member 9 | |||
| calcium | Cre06.g263950.t1.2 | Cre06.g263950.t1.3 | Na+/K + ATPase, alpha subunit | ||
| metabolite transporters at the envelope membrane | Cre08.g363600.t1.1 | Cre08.g363600.t1.2 | Glucose-6-phosphate, PEP/phosphate antiporter | ||
| metal | Cre17.g720400.t1.2 | Cre17.g720400.t1.3 | HMA1 | Heavy metal transporting ATPase | |
| P- and V-ATPases | Cre10.g459200.t1.1 | Cre10.g459200.t1.2 | ACA4 | Plasma membrane H + -transporting ATPase | |
| phosphate | Cre02.g144650.t1.1 | Cre02.g144650.t1.2 | PTB12 | Na+/Pi symporter | |
| potassium | Cre06.g278700.t1.2 | Cre06.g278700.t1.2 | Myotrophin and similar proteins | ||
-
*
Functional terms are inferred by homology to the annotation set of Arabidopsis thaliana (Lopez et al., 2011).
-
†
Corresponding gene model was not found in v5.
-
‡
No functional annotations found on v5 but defined by MapMan on Algal Functional Annotation Tool (Lopez et al., 2011).
-
§
Induction during 1O2 acclimation dependent on SAK1 (Table 5).
1O2 response genes are not induced when RB is added in the dark
| Fold change +RB/−RB (SD)* | ||
|---|---|---|
| Gene name or ID | WT | sak1 |
| GPX5 | 1.13 (0.33) | 0.87 (0.31) |
| SAK1 | 1.38 (0.08) | 1.29 (0.19) |
| CFA1 | 0.90 (0.04) | 1.44 (0.22) |
| SOUL2 | 1.17 (0.25) | 1.11 (0.19) |
| MRP3†,‡ | 1.13 (0.12) | 1.07 (0.25) |
| Cre12.g503950†,‡ | 0.93 (0.06) | 1.20 (0.12) |
| Cre14.g613950†,§ | 0.65 (0.06) | 0.79 (0.15) |
| Cre04.g220850†,‡ | 1.00 (0.09) | 1.29 (0.04) |
| Cre09.g395750†,‡ | 1.05 (0.10) | 1.29 (0.12) |
-
*
Average of fold change and standard deviation (SD) of biological triplicates.
-
†
Annotated as transport function.
-
‡
ABC transporter.
-
§
Sec14-like phosphatidylinositol transfer protein.
Genes that require SAK1 for induction by 1O2
| Gene ID (v4) | Gene ID (v5) | Gene name | Annotation | FC WT* (log2) | FC sak1 (log2) | Attenuation (FC-sak1/FC-WT)† | Basal repression in sak1 (log2) |
|---|---|---|---|---|---|---|---|
| Cre02.g137700.t1.1‡ | Cre09.g400404 | 6.49 | 1.80 | 0.04 | −3.35 | ||
| Cre06.g281250.t1.1‡ | Cre06.g281250 | CFA1 | Cyclopropane fatty acid synthase | 5.92 | 1.16 | 0.04 | −2.10 |
| Cre27.g775950.t1.2 | Cre12.g557928 | 5.83 | 0.81 | 0.03 | |||
| Cre01.g033300.t1.1 | Cre01.g033300 | 5.72 | −0.39 | 0.01 | |||
| Cre13.g566850.t1.1‡ | Cre13.g566850 | SOUL2 | SOUL heme-binding protein | 5.53 | 1.33 | 0.05 | −2.60 |
| Cre14.g623650.t1.1 | Cre14.g623650 | Alcohol dehydrogenase | 4.89 | 1.67 | 0.11 | ||
| Cre13.g600650.t1.1 | Cre06.g278245 | Rieske 2Fe-2S domain | 4.76 | 1.64 | 0.12 | ||
| Cre06.g263550.t1.1 | Cre06.g263550 | LCI7 | R53.5-related protein | 4.46 | 1.77 | 0.15 | |
| Cre07.g342100.t1.1 | Cre07.g342100 | 4.43 | 1.40 | 0.12 | |||
| Cre06.g299700.t1.1‡ | Cre06.g299700 | SOUL1 | SOUL heme-binding protein | 4.32 | 0.43 | 0.07 | −1.13 |
| Cre09.g398700.t1.1‡ | Cre09.g398700 | CFA2 | Cyclopropane fatty acid synthase | 4.05 | 0.18 | 0.07 | −1.00 |
| Cre12.g492650.t1.1‡ | Cre12.g492650 | FAS2 | Fasciclin-like protein | 4.01 | 0.07 | 0.07 | −1.24 |
| Cre08.g381510.t1.1 | NF | 3.94 | 0.73 | 0.11 | |||
| Cre10.g458450.t1.2 | Cre10.g458450 | GPX5 | Glutathione peroxidase | 3.91 | 2.06 | 0.28 | |
| Cre11.g474600.t1.1 | Cre02.g095151 | ABC transporter (ABC-2 type) | 3.90 | 0.44 | 0.09 | ||
| Cre13.g600700.t1.1 | Cre06.g278246 | 3.78 | 1.48 | 0.20 | |||
| Cre14.g613950.t1.1 | Cre14.g613950 | 3.65 | 1.38 | 0.21 | |||
| Cre06.g269300.t1.1 | Cre06.g269300 | DUF1365 | 3.50 | 0.40 | 0.12 | ||
| Cre08.g380300.t1.2 | Cre08.g380300 | MSRA3 | Peptide methionine sulfoxide reductase | 3.45 | 0.66 | 0.14 | |
| Cre28.g776450.t1.1 | Cre08.g358573 | TRP7 | Transient receptor potential ion channel | 3.31 | −0.79 | 0.06 | |
| Cre01.g031650.t1.2 | Cre01.g031650 | CGLD12 | Potential galactosyl transferase activity | 3.30 | 0.67 | 0.16 | |
| Cre14.g629061.t1.1 | NF | DUF2177 | 3.25 | 0.08 | 0.11 | ||
| Cre12.g503950.t1.1 | Cre12.g503950 | CRAL/TRIO domain | 3.24 | 0.31 | 0.13 | ||
| Cre13.g564900.t1.1 | Cre13.g564900 | ABC transporter transmembrane region | 3.22 | 0.34 | 0.14 | ||
| Cre02.g139500.t1.1 | Cre09.g401701 | DUF1295 | 3.04 | −0.16 | 0.11 | ||
| Cre14.g618400.t1.1 | Cre14.g618400 | 2.97 | 1.15 | 0.28 | |||
| Cre17.g715150.t1.1 | Cre17.g715150 | 2.89 | 0.13 | 0.15 | |||
| Cre17.g741300.t1.2‡ | Cre17.g741300 | SAK1 | 2.88 | 0.66 | 0.21 | −2.77 | |
| Cre01.g007300.t1.1 | Cre01.g007300 | 2.85 | −1.15 | 0.06 | |||
| Cre16.g648700.t1.2‡ | Cre16.g648700 | ABC transporter (ABC-2 type) | 2.79 | 0.26 | 0.17 | −1.26 | |
| Cre13.g566900.t1.2 | Cre13.g566900 | 2.76 | −0.38 | 0.11 | |||
| Cre02.g137750.t1.2 | Cre09.g400441 | JmjC domain | 2.72 | −0.31 | 0.12 | ||
| Cre06.g263500.t1.1 | Cre06.g263500 | Archease protein family (DUF101) | 2.67 | 1.02 | 0.32 | ||
| Cre01.g016150.t1.1‡ | Cre01.g016150 | ADP-ribosylglycohydrolase | 2.65 | 0.17 | 0.18 | −1.26 | |
| Cre08.g380000.t1.1 | Cre08.g380000 | Formylglycine-generating sulfatase enzyme | 2.59 | 1.53 | 0.48 | ||
| Cre14.g615600.t1.1 | Cre14.g615600 | Putative serine esterase (DUF676) | 2.53 | −0.54 | 0.12 | ||
| Cre11.g472900.t1.2 | Cre02.g095113 | CAP-Gly domain | 2.45 | −0.05 | 0.18 | ||
| Cre06.g269250.t1.1 | Cre06.g269250 | 2.44 | 0.55 | 0.27 | |||
| Cre02.g120600.t1.1 | Cre09.g403071 | 2.44 | 0.94 | 0.35 | |||
| Cre06.g261200.t1.1 | Cre06.g261200 | ERG25 | Sterol desaturase | 2.42 | 0.64 | 0.29 | |
| Cre16.g683400.t1.1 | Cre16.g683400 | CRAL/TRIO domain | 2.40 | 0.08 | 0.20 | ||
| Cre22.g765150.t1.1 | Cre11.g467725 | hypothetical protein | 2.30 | 0.46 | 0.28 | ||
| Cre13.g571800.t1.2 | Cre13.g571800 | DUF1336 | 2.27 | 0.72 | 0.34 | ||
| Cre13.g579450.t1.2 | Cre13.g579450 | CST1 | Membrane transporter | 2.27 | 1.23 | 0.49 | |
| Cre08.g380350.t1.1 | Cre08.g380350 | 2.21 | −0.01 | 0.21 | |||
| Cre16.g649250.t1.2 | Cre16.g649250 | 2.08 | 0.58 | 0.35 | |||
| Cre11.g476250.t1.1 | Cre11.g476250 | 2.08 | 0.49 | 0.33 | |||
| Cre02.g108000.t1.2 | Cre02.g108000 | 2.08 | 1.03 | 0.49 | |||
| Cre13.g583300.t1.1 | Cre13.g583300 | 1.98 | −0.48 | 0.18 | |||
| Cre04.g215300.t1.2 | NF | 1.97 | 0.57 | 0.38 | |||
| Cre02.g139450.t1.1 | Cre09.g401663 | DUF947 | 1.95 | −0.62 | 0.17 | ||
| Cre03.g194750.t1.2 | Cre03.g194750 | 1.95 | 0.73 | 0.43 | |||
| Cre06.g258600.t1.1 | Cre06.g258600 | Dienelactone hydrolase family | 1.91 | −0.95 | 0.14 | ||
| Cre10.g418700.t1.1 | Cre10.g418700 | Probable N6-adenine methyltransferase | 1.87 | −0.03 | 0.27 | ||
| Cre10.g444550.t1.1 | Cre10.g444550 | SPP1A | Signal peptide peptidase | 1.81 | 0.51 | 0.41 | |
| Cre01.g060050.t1.2 | Cre03.g145807 | 1.78 | −0.11 | 0.27 | |||
| Cre09.g410050.t1.1 | Cre09.g410050 | Calcium transporting ATPase | 1.76 | 0.51 | 0.42 | ||
| Cre03.g163400.t1.2 | Cre03.g163400 | 1.76 | −0.17 | 0.26 | |||
| Cre01.g008450.t1.1 | Cre01.g008450 | Nuf2 family | 1.73 | −0.54 | 0.21 | ||
| Cre12.g536650.t1.1 | Cre12.g536650 | 1.72 | 0.35 | 0.39 | |||
| Cre02.g114900.t1.2 | Cre02.g114900 | ANK23 | predicted protein | 1.71 | 0.08 | 0.32 | |
| Cre16.g661850.t1.2 | Cre16.g661850 | Calcium/calmoduline dependent protein kinase association | 1.69 | 0.03 | 0.32 | ||
| Cre14.g615500.t1.2 | Cre14.g615500 | Glycoprotease family | 1.68 | −0.76 | 0.18 | ||
| Cre11.g483100.t1.2 | Cre11.g483100 | Protein kinase | 1.66 | −0.49 | 0.22 | ||
| Cre28.g776650.t1.1 | Cre08.g358569 | 1.64 | 0.33 | 0.40 | |||
| Cre07.g340250.t1.2 | Cre07.g340250 | Protein kinase | 1.63 | −0.41 | 0.24 | ||
| Cre06.g296250.t1.2 | Cre06.g296250 | SYK1 | tRNA synthetase, class II | 1.60 | 0.54 | 0.48 | |
| Cre06.g310500.t1.1 | Cre06.g310500 | 1.57 | 0.18 | 0.38 | |||
| Cre07.g342800.t1.2 | Cre07.g342800 | CGL16 | Predicted protein | 1.49 | 0.32 | 0.44 | |
| Cre03.g181450.t1.2 | Cre03.g181450 | DUF1619 | 1.47 | 0.35 | 0.46 | ||
| Cre66.g793601.t1.1 | Cre35.g759497 | 1.47 | 0.03 | 0.37 | |||
| Cre14.g614050.t1.2 | Cre14.g614050 | MAP65 | Microtubule associated protein | 1.43 | 0.06 | 0.39 | |
| Cre04.g217500.t1.1 | Cre04.g217500 | Inosine-uridine preferring nucleoside hydrolase | 1.42 | 0.19 | 0.43 | ||
| Cre06.g292950.t1.1 | Cre06.g292950 | DNA polymerase delta, subunit 4 | 1.38 | −0.12 | 0.35 | ||
| Cre16.g661750.t1.1 | Cre16.g661750 | Calcium/calmoduline dependent protein kinase association | 1.38 | −0.12 | 0.35 | ||
| Cre01.g007000.t1.1 | Cre01.g007000 | ABC transporter (ABC-2 type) | 1.35 | 0.21 | 0.45 | ||
| Cre04.g224400.t1.2 | Cre04.g224400 | ABC transporter (ABC-2 type) | 1.34 | −0.13 | 0.36 | ||
| Cre01.g068400.t1.2 | Cre16.g680790 | 1.33 | 0.16 | 0.45 | |||
| Cre05.g237400.t1.1 | Cre05.g237400 | DAE1 | Diaminopimelate epimerase | 1.32 | 0.22 | 0.47 | |
| Cre14.g609600.t1.2 | Cre14.g609600 | 1.32 | −0.58 | 0.27 | |||
| Cre05.g234850.t1.2 | Cre05.g234850 | Ubiquitin carboxyl-terminal hydrolase | 1.29 | 0.16 | 0.46 | ||
| Cre03.g179200.t1.1 | Cre03.g179200 | 1.28 | −0.48 | 0.30 | |||
| Cre10.g417730.t1.1 | Cre10.g417730 | 1.27 | 0.17 | 0.47 | |||
| Cre03.g159700.t1.2 | Cre03.g159700 | 1.26 | −0.14 | 0.38 | |||
| Cre12.g540150.t1.2 | Cre12.g540150 | 1.19 | −0.24 | 0.37 | |||
| Cre01.g006550.t1.2‡ | Cre01.g006550 | No annotation | 1.17 | −0.49 | 0.32 | −1.60 | |
| Cre03.g159950.t1.2 | Cre03.g159950 | 1.17 | −0.17 | 0.40 | |||
| Cre27.g775900.t1.2 | Cre12.g557503 | 1.14 | −0.70 | 0.28 | |||
| Cre02.g121600.t1.1 | Cre09.g387208 | Protein kinase | 1.14 | 0.00 | 0.46 | ||
| Cre14.g609550.t1.1 | NF | 1.13 | −0.84 | 0.26 | |||
| Cre07.g315050.t1.2 | Cre07.g315050 | 1.12 | −0.03 | 0.45 | |||
| Cre04.g218800.t1.2 | Cre04.g218800 | THB3 | Truncated hemoglobin | 1.11 | −0.50 | 0.33 | |
| Cre02.g133300.t1.1 | Cre09.g396624 | 1.11 | −0.43 | 0.34 | |||
| Cre01.g060650.t1.2 | Cre03.g146067 | 1.10 | −0.42 | 0.35 | |||
| Cre01.g057050.t1.1 | Cre03.g144324 | 1.10 | 0.04 | 0.48 | |||
| Cre06.g304950.t1.1 | Cre06.g304950 | 1.07 | −0.65 | 0.30 | |||
| Cre08.g358200.t1.2 | Cre08.g358200 | A4 | Protein kinase | 1.07 | −0.82 | 0.27 | |
| Cre16.g689550.t1.2 | Cre16.g689550 | PTK8 | Putative tyrosine kinase | 1.06 | −0.17 | 0.43 | |
| Cre17.g720950.t1.1 | Cre17.g720950 | 3-oxo-5-alpha-steroid 4-dehydrogenase | 1.05 | −0.26 | 0.40 | ||
| Cre02.g090950.t1.2 | Cre02.g090950 | 1.05 | −0.27 | 0.40 | |||
| Cre16.g683350.t1.1 | Cre16.g683350 | 1.03 | −0.67 | 0.31 | |||
| Cre02.g109450.t1.1 | Cre02.g109450 | 1.01 | −0.03 | 0.48 | |||
| Cre16.g652750.t1.1 | Cre16.g652750 | 1.01 | −0.29 | 0.41 | |||
| Cre03.g190000.t1.1 | Cre03.g190000 | 1.00 | −0.99 | 0.25 |
-
*
Data were ordered by FC in WT.
-
†
Of the 52 most highly induced genes in WT (the top 10%), 37 were SAK1-dependent, and the induction of 33 of these genes was strongly attenuated to only 0.01-0.25 of magnitude of FC found in the WT. Dashed line indicates cutoff of FC for the top 10% most strongly induced genes.
-
‡
Genes that are repressed at basal level in sak1.
-
NF, not found in v5.
Enriched functional classes among differentially expressed genes in sak1 during 1O2 acclimation
| Primary Mapman class | Secondary Mapman class | Gene ID (v4) | Gene name | Annotation |
|---|---|---|---|---|
| Up-regulated genes | ||||
| Secondary metabolism | isoprenoids | Cre13.g565650.t1.1 | Geranylgeranyl pyrophosphate synthase/Polyprenyl synthetase | |
| Cre06.g267600.t1.1 | Lycopene epsilon cyclase | |||
| Cre09.g407200.t1.1 | Phytoene desaturase | |||
| Cre06.g267600.t1.1 | Lycopene epsilon cyclase | |||
| Cre01.g011100.t1.1 | Prenyltransferase and squalene oxidase repeat, Oxidosqualene-lanosterol cyclase and related proteins | |||
| N misc | Cre08.g381707.t1.1 | NF* | ||
| phenylpropanoids | Cre03.g207800.t1.1 | Alcohol dehydrogenase, class V | ||
| Cre14.g623650.t1.1 | Alcohol dehydrogenase, class V (Zinc-binding) | |||
| Cre01.g039350.t1.1 | Cytochrome P450 reductase, possibly CYP505B family | |||
| sulfur-containing | Cre06.g299400.t1.1 | NF* | ||
| wax | Cre17.g722150.t1.1 | PKS3 | Type III polyketide synthase | |
| Cre07.g318500.t1.2 | FAE1/Type III polyketide synthase-like protein, Chalcone and stilbene synthases | |||
| Lipid metabolism | ‘exotics’ (steroids, squalene etc) | Cre01.g061750.t1.1 | serine palmitoyltransferase | |
| Cre02.g137850.t1.1 | NF* | |||
| Cre83.g796250.t1.1 | NF* | |||
| Cre01.g011100.t1.1 | Prenyltransferase and squalene oxidase repeat, Oxidosqualene-lanosterol cyclase and related proteins | |||
| FA synthesis and FA elongation | Cre06.g256750.t1.1 | Acyl carrier protein thioesterase | ||
| Cre03.g182050.t1.1 | Long-chain acyl-CoA synthetases (AMP-forming) | |||
| Cre02.g074650.t1.1 | Kelch repeat-containing proteins, Acyl-CoA binding protei | |||
| glycerol metabolism | Cre01.g053000.t1.1 | GPD2 | Glycerol-3-phosphate dehydrogenase/dihydroxyacetone-3-phosphate reductase | |
| glycolipid synthesis | Cre13.g583600.t1.1 | DGD1 | Digalactosyldiacylglycerol synthase | |
| lipid degradation | Cre01.g057450.t1.2 | NF* | ||
| Cre02.g126050.t1.1 | NF* | |||
| phospholipid synthesis | Cre06.g281250.t1.1 | CFA1 | Cyclopropane fatty acid synthase | |
| Cre01.g038250.t1.1 | SDC1 | Serine decarboxylase | ||
| Cre11.g472700.t1.1 | NF* | |||
| Cre13.g604700.t1.2 | CDP-alcohol phosphatidyltransferase/Phosphatidylglycerol-phosphate synthase | |||
| Cell | vesicle transport | Cre18.g744100.t1.1 | NF* | |
| Cre17.g721900.t1.1 | COG5 | Component of oligomeric golgi complex | ||
| Cre01.g003050.t1.1 | SEC8 | Component of the Exocyst Complex | ||
| Cre04.g224800.t1.1 | Endosomal R-SNARE protein, Vamp7/Nyv1-family | |||
| Cre17.g728150.t1.1 | Endosomal R-SNARE protein, Yky6-family | |||
| Cre12.g507450.t1.1 | Trans-Golgi network Qa-SNARE protein, Syntaxin16/Syx16/Tlg2/Syp4-family | |||
| Cre03.g210600.t1.1 | NF* | |||
| Cre04.g225900.t1.1 | Endosomal R-SNARE protein, Vamp7/Nyv1-family | |||
| Cre02.g101400.t1.1 | CHC1 | Clathrin Heavy Chain | ||
| Cre17.g709350.t1.1 | Late endosomal Qc-SNARE protein, Syx8/Syntaxin8-family | |||
| Cre07.g342050.t1.1 | Endosomal Qb-SNARE, Npsn-family | |||
| Cre16.g692050.t1.1 | ER-Golgi Qa-SNARE protein, Syntaxin5/Syx5/Sed5/Syp3-family | |||
| Cre16.g676650.t1.1 | AP1G1 | Gamma1-Adaptin | ||
| Cre02.g099000.t1.1 | Late endosomal Qc-SNARE protein, Syx6/Tlg1/Syp5/6-family | |||
| Cre12.g554200.t1.2 | ER-Golgi Qb-SNARE, Memb/GS35/Bos1-family | |||
| Cre06.g310000.t1.1 | AP4E1 | Epsilon4-Adaptin | ||
| Cre10.g421250.t1.1 | EXO70 | Hypothetical Conserved Protein. Similar to Exo70, a subunit of the exocyst complex | ||
| Cre07.g330950.t1.1 | AP4S4 | Sigma4-Adaptin | ||
| Cre12.g488850.t1.2 | Adaptin, alpha/gamma/epsilon | |||
| division | Cre06.g269950.t1.1 | CDC48 | Protein involved in ubiquitin-dependent degradation of ER-bound substrates | |
| Cre08.g359200.t1.2 | Regulator of chromosome condensation (RCC1) | |||
| organisation | Cre13.g588600.t1.2 | Kinesin (SMY1 subfamily) | ||
| Cre12.g513450.t1.1 | TUH1 | Eta-Tubulin | ||
| Cre01.g010950.t1.2 | 26S proteasome regulatory complex, subunit PSMD10 (Ankyrin repeat) | |||
| Cre16.g679650.t1.2 | Fimbrin/Plastin | |||
| Cre06.g261950.t1.1 | Myotrophin and similar proteins (Ankyrin repeat) | |||
| Cre06.g291700.t1.1 | RSP3 | Radial spoke protein 3 | ||
| Cre10.g446700.t1.1 | ANK28 | Ankyrin repeat and DHHC-type Zn-finger domain containing proteins | ||
| Hormone metabolism† | abscisic acid | Cre16.g657800.t1.2 | CCD3 | Carotenoid cleavage dioxygenase |
| auxin | Cre14.g609900.t1.1 | Predicted membrane protein, contains DoH and Cytochrome b-561/ferric reductase transmembrane domains | ||
| brassinosteroid | Cre16.g663950.t1.1 | Sterol C5 desaturase | ||
| Cre02.g092350.t1.1 | Cytochrome P450, CYP51 superfamily; sterol 14 desaturase | |||
| Cre12.g557900.t1.1 | CDI1 | C-8,7 sterol isomerase | ||
| Cre02.g076800.t1.1 | Delta14-sterol reductase, mitochondrial | |||
| Cre12.g500500.t1.2 | 24-methylenesterol C-methyltransferase | |||
| ethylene | Cre02.g108450.t1.1 | FAP280 | Flagellar Associated Protein, transcriptional coactivator-like, putative transcription factor | |
| jasmonate | Cre19.g756100.t1.1 | NF* | ||
| Misc | acid and other phosphatases | Cre09.g396900.t1.1 | NADH pyrophosphatase I of the Nudix family of hydrolases | |
| Cre06.g259650.t1.1 | Calcineurin-like phosphoesterase, Acid-phosphatase-related | |||
| Cre06.g249800.t1.1 | Sphingomyelin synthetase -related | |||
| cytochrome P450 | Cre05.g234100.t1.1 | Cytochrome P450, CYP197 superfamily | ||
| dynamin | Cre02.g079550.t1.1 | DRP2 | Dynamin-related GTPase, involved in circadian rhythms | |
| Cre05.g245950.t1.1 | DRP1 | Dynamin-related GTPase | ||
| glutathione S transferases | Cre03.g154950.t1.1 | Glutathione S-transferase | ||
| misc2 | Cre12.g538450.t1.1 | EPT1 | CDP-Etn:DAG Ethanolamine phosphotransferase | |
| short chain dehydrogenase/reductase (SDR) | Cre12.g556750.t1.2 | Short-chain dehydrogenase/reductase | ||
| Cre08.g384864.t1.1 | SH3 domain, protein binding | |||
| Cre27.g775000.t1.1 | NF* | |||
| Cre17.g731350.t1.2 | Short chain dehydrogenase | |||
| UDP glucosyl and glucoronyl transferases | Cre02.g111150.t1.2 | ELG26 | Exostosin-like glycosyltransferase | |
| Cre02.g144050.t1.1 | Acetylglucosaminyltransferase EXT1/exostosin 1 | |||
| Cre03.g204050.t1.2 | ELG6 | Exostosin-like glycosyltransferases | ||
| Cre11.g474450.t1.1 | NF* | |||
| Cre03.g173300.t1.1 | Lactosylceramide 4-alpha-galactosyltransferase (alpha- 1,4-galactosyltransferase) | |||
| Cre02.g116600.t1.1 | ELG23 | Exostosin-like glycosyltransferase | ||
| Down-regulated genes | ||||
| Hormone metabolism† | cytokinin | Cre18.g744950.t1.2 | NF* | |
| Cre16.g678900.t1.1 | Response regulator receiver domain | |||
| Cre01.g040450.t1.1 | HDT1 | Histidine-aspartic acid phosphotransferase 1 (phosphorylation cascade) | ||
| ethylene | Cre09.g403550.t1.1 | Iron/ascorbate family oxidoreductases | ||
| Nucleotide metabolism | deoxynucleotide metabolism | Cre12.g491050.t1.1 | RIR2 | Ribonucleotide reductase (RNR), small subunit |
| Cre12.g492950.t1.1 | RIR1 | Ribonucleotide reductase (RNR), large subunit, class I | ||
| Cre16.g667850.t1.1 | dUTP pyrophosphatase | |||
| synthesis | Cre14.g614300.t1.1 | Inosine-5-monophosphate dehydrogenase/GMP reductase | ||
| Cre07.g318750.t1.1 | Phosphoribosylformylglycinamidine cyclo-ligase | |||
| Tetrapyrrole synthesis | porphobilinogen deaminase | Cre16.g663900.t1.1 | Porphobilinogen deaminase | |
| protochlorophyllide reductase | Cre01.g015350.t1.1 | Light-dependent protochlorophyllide reductase | ||
| urogen III methylase | Cre02.g133050.t1.2 | NF* | ||
| DNA | repair | Cre16.g670550.t1.2 | XP-G/RAD2 DNA repair endonuclease | |
| synthesis/chromatin structure | Cre07.g338000.t1.1 | MCM2 | Minichromosome maintenance protein | |
| Cre07.g314900.t1.2 | ATP-dependent RNA helicase, DEAD/DEAH helicase | |||
| Cre03.g172950.t1.1 | CBF5 | Centromere/microtubule binding protein | ||
| Cre01.g015250.t1.1 | Eukaryotic DNA polymerase delta | |||
| Cre27.g774200.t1.2 | NF* | |||
| Cre07.g316850.t1.1 | MCM4 | Minichromosome maintenance protein | ||
| unspecified | Cre10.g451250.t1.2 | Adenylate and guanylate cyclase catalytic domain, 3-5 exonuclease | ||
| Cre01.g059950.t1.2 | NF* | |||
-
*
Corresponding gene model was not found in v5.
-
†
Functional terms are inferred by homology to the annotation set of Arabidopsis thaliana (Lopez et al., 2011).
Enriched functional classes among differentially expressed genes in sak1 at basal level
| Primary Mapman class | Secondary Mapman class | Gene ID (v4) | Gene name | Annotation |
|---|---|---|---|---|
| Elevated in sak1 | ||||
| nucleotide metabolism | deoxynucleotide metabolism | Cre12.g491050.t1.1 | RIR2 | Ribonucleotide reductase (RNR), small subunit |
| Cre12.g492950.t1.1 | RIR1 | Ribonucleotide reductase (RNR), large subunit, class I | ||
| Cre16.g667850.t1.1 | dUTP pyrophosphatase | |||
| phosphotransfer and pyrophosphatases | Cre02.g122450.t1.1 | NF* | ||
| Cre02.g093950.t1.1 | PYR5 | Uridine 5'- monophosphate synthase/orotate phosphoribosyltransferase | ||
| Cre12.g519950.t1.1 | Flagellar Associated Protein similar to adenylate/guanylate kinases | |||
| Cre26.g772450.t1.1 | NF* | |||
| synthesis | Cre65.g793400.t1.1 | NF* | ||
| Cre02.g079700.t1.1 | PYR2 | Aspartate carbamoyltransferase | ||
| Cre01.g048950.t1.1 | dUTP pyrophosphatase | |||
| Cre07.g318750.t1.1 | Phosphoribosylformylglycinamidine cyclo-ligase. | |||
| DNA | repair | Cre07.g314650.t1.1 | Chloroplast RecA recombination protein | |
| synthesis/chromatin structure | Cre04.g214350.t1.2 | Eukaryotic DNA polymerase alpha, catalytic subunit | ||
| Cre07.g314900.t1.2 | ATP-dependent RNA helicase (DEAD/DEAH) | |||
| Cre04.g223850.t1.1 | Cytoplasmic DExD/H-box RNA helicase | |||
| Cre01.g015250.t1.1 | Eukaryotic DNA polymerase delta, catalytic subunit. | |||
| Cre07.g342506.t1.1 | Ubiquitin-protein ligase | |||
| Cre07.g338000.t1.1 | MCM2 | Minichromosome maintenance protein | ||
| Cre03.g178650.t1.1 | MCM6 | MCM6 DNA replication protein | ||
| Cre07.g312350.t1.2 | DNA polymerase alpha, primase subunit | |||
| Cre01.g009250.t1.2 | TOP2 | DNA topoisomerase II | ||
| Cre26.g772150.t1.1 | NF* | |||
| Cre07.g316850.t1.1 | MCM4 | Minichromosome maintenance protein 4 | ||
| Cre06.g263800.t1.2 | tRNA-splicing endonuclease positive effector (SEN1) | |||
| Cre06.g295700.t1.2 | MCM3 | Minichromosome maintenance protein | ||
| Cre06.g251800.t1.1 | RFC4 | DNA replication factor C complex subunit 4 | ||
| unspecified | Cre07.g322300.t1.2 | DNA repair helicase of the DEAD superfamily | ||
| Cre17.g718100.t1.1 | Phosphatidylinositol transfer protein SEC14 and related proteins (CRAL/TRIO) | |||
| Tetrapyrrole synthesis | Glu-tRNA reductase | Cre07.g342150.t1.1 | HEM1 | Glutamyl-tRNA reductase |
| Glu-tRNA synthetase | Cre44.g788000.t1.1 | Glutamyl-tRNA reductase | ||
| Cre06.g306300.t1.1 | CHLI1 | Magnesium chelatase subunit I | ||
| magnesium chelatase | Cre07.g325500.t1.1 | Magnesium chelatase subunit H | ||
| protochlorophyllide reductase | Cre01.g015350.t1.1 | POR1 | Light-dependent protochlorophyllide reductase | |
| Photosynthesis | Calvin-Benson cycle | Cre05.g234550.t1.1 | Fructose-biphosphate aldolase | |
| light reaction | Cre07.g330250.t1.1 | PSAH | Subunit H of photosystem I | |
| Cre07.g334550.t1.1 | Photosystem I subunit PsaO | |||
| Cre06.g261000.t1.1 | PSBR | 10 kDa photosystem II polypeptide | ||
| photorespiration | Cre12.g542300.t1.1 | GYK1 | Glycerate kinase | |
| Cre06.g253350.t1.1 | GCSH | Glycine cleavage system, H-protein | ||
| Cre06.g293950.t1.1 | SHMT2 | Serine hydroxymethyltransferase 2 | ||
| Transport | ABC transporters and multidrug resistance systems | Cre04.g222700.t1.1 | ATPase component of ABC transporters with duplicated ATPase domains/Translation elongation factor EF-3b | |
| Cre17.g728400.t1.2 | ABCtransporter (ABC-2 type) | |||
| Cre05.g241350.t1.2 | ABCtransporter (ABC-2 type) | |||
| Cre03.g169300.t1.1 | ABCtransporter (ABC-2 type) | |||
| Cre11.g474600.t1.1 | NF* | |||
| amino acids | Cre04.g226150.t1.2 | AOC1 | Amino acid carrier 1; belongs to APC (Amino acid Polyamine organo Cation) family | |
| calcium | Cre09.g388850.t1.1 | ACA1 | P-type ATPase/cation transporter, plasma membrane | |
| metabolite transporters at the envelope membrane | Cre06.g263850.t1.2 | TPT2 | Triose phosphate/phosphate translocator | |
| metabolite transporters at the mitochondrial membrane | Cre10.g449100.t1.1 | Mitochondrial oxodicarboxylate carrier protein | ||
| Cre01.g069350.t1.1 | NF* | |||
| Cre15.g641200.t1.1 | Mitochondrial fatty acid anion carrier protein/Uncoupling protein | |||
| Cre09.g396350.t1.1 | Mitochondrial carrier protein PET8 | |||
| misc | Cre06.g311000.t1.2 | FBT2 | Folate transporte | |
| Cre17.g718100.t1.1 | Phosphatidylinositol transfer protein SEC14 and related proteins (CRAL/TRIO) | |||
| phosphate | Cre16.g686750.t1.1 | PTA3 | Proton/phosphate symporter | |
| Cre16.g675300.t1.2 | Sodium-dependent phosphate transporter, major facilitator superfamily | |||
| potassium | Cre12.g553450.t1.2 | NF* | ||
| sulphate | Cre17.g723350.t1.1 | SUL2 | Sulfate anion transporter | |
| unspecified cations | Cre13.g573900.t1.1 | Na+:iodide/myo-inositol/multivitamin symporters | ||
| sugars | Cre16.g675300.t1.2 | Sodium-dependent phosphate transporter, major facilitator superfamily | ||
| RNA | processing | Cre10.g427700.t1.1 | ATP-dependent RNA helicase, DEAD/DEAH box helicase | |
| Cre12.g538750.t1.1 | LSM1 | U6 snRNA-associated Sm-like protein LSm1, RNA cap binding; (SMP6d) | ||
| Cre10.g433750.t1.2 | PAP1 | Nuclear poly(A) polymerase | ||
| Cre03.g182950.t1.1 | NF* | |||
| Cre08.g375128.t1.1 | NF* | |||
| regulation of transcription | Cre17.g728200.t1.2 | YL-1 protein (transcription factor-like 1) | ||
| Cre06.g275500.t1.1 | AP2 Transcription factor | |||
| Cre28.g777500.t1.2 | NF* | |||
| Cre13.g572450.t1.1 | Response regulator receiver domain (sensor histidine kinase-related, regulation of transcription) | |||
| Cre14.g620500.t1.1 | AP2 Transcription factor | |||
| Cre16.g673150.t1.1 | Histone deacetylase complex, catalytic component RPD3 | |||
| Cre02.g078700.t1.2 | DNA damage-responsive repressor GIS1/RPH1, jumonji superfamily | |||
| Cre03.g198800.t1.1 | Myb-like DNA-binding domain | |||
| Cre04.g218050.t1.2 | RWP-RK domain | |||
| Cre07.g324400.t1.1 | VPS24 | Subunit of the ESCRT-III complex, vaculoar sortin protein | ||
| Cre11.g481050.t1.1 | SWI/SNF-related chromatin binding protein | |||
| Cre02.g101950.t1.1 | TMU2 | tRNA (uracil-5)-methyltransferase | ||
| Cre10.g459600.t1.2 | CAATT-binding transcription factor/60S ribosomal subunit biogenesis protein | |||
| Cre01.g018650.t1.2 | NF* | |||
| Cre01.g012200.t1.2 | NF* | |||
| Cre02.g129750.t1.1 | NF* | |||
| Cre10.g461750.t1.2 | DNA (cytosine-5-)-methyltransferase | |||
| Cre01.g004600.t1.2 | RWP12 | Putative RWP-RK domain transcription factor | ||
| Cre09.g400100.t1.1 | Predicted Zn-finger protein, zinc and DNA binding domains | |||
| Cre07.g335150.t1.2 | SBP domain | |||
| RNA binding | Cre16.g662700.t1.1 | NF* | ||
| Cre07.g330300.t1.1 | RNA-binding protein musashi/mRNA cleavage and polyadenylation factor I complex, subunit HRP1 | |||
| Cre06.g275100.t1.1 | RNA-binding protein musashi/mRNA cleavage and polyadenylation factor I complex, subunit HRP1 | |||
| transcription | Cre07.g322200.t1.1 | NF* | ||
| Repressed in sak1 | ||||
| Transport | ABC transporters and multidrug resistance systems | Cre02.g097800.t1.2 | ABC transporter (MDR) | |
| Cre17.g725200.t1.1 | ABC transporter, peptide exporter | |||
| Cre13.g580300.t1.1 | ABC transporter family protein | |||
| Cre10.g439000.t1.2 | Long-chain acyl-CoA transporter, ABC superfamily (involved in peroxisome organization and biogenesis) | |||
| amino acids | Cre06.g292350.t1.1 | AOC4 | Amino acid carrier | |
| calcium | Cre06.g263950.t1.2 | Sodium/potassium-transporting ATPase subunit alpha | ||
| Cre16.g681750.t1.2 | Calcium transporting ATPase | |||
| metabolite transporters at the mitochondrial membrane | Cre03.g172300.t1.1 | Mitochondrial phosphate carrier protein | ||
| Cre09.g394800.t1.2 | Mitochondrial substrate carrier protein | |||
| metal | Cre03.g189550.t1.2 | ZIP3 | Zinc transporter, ZIP family | |
| Cre11.g479600.t1.2 | Sodium/calcium exchanger NCX1 and related proteins | |||
| Cre06.g281900.t1.1 | ZIP7 | Zinc transporter and related ZIP domain-containing proteins | ||
| misc | Cre02.g089900.t1.1 | Secretory carrier membrane protein | ||
| Cre10.g448050.t1.1 | Retinaldehyde binding protein-related (CRAL/TRIO domain) | |||
| Cre03.g177750.t1.2 | Multidrug resistance pump | |||
| NDP-sugars at the ER | Cre02.g112900.t1.1 | GDP-fucose transporter (Triose-phosphate transporter family) | ||
| P- and V-ATPases | Cre01.g027800.t1.1 | ATPvH | Vacuolar ATP synthase subunit H | |
| Cre10.g446550.t1.1 | ATPvF | Vacuolar ATP synthase subunit F | ||
| Cre03.g176250.t1.1 | ATPvD1 | Vacuolar ATP synthase subunit D | ||
| Cre06.g250250.t1.1 | ATPvC | Vacuolar ATP synthase subunit C | ||
| Cre10.g459200.t1.1 | ACA4 | P-type ATPase/cation transporter, plasma membrane (Low CO2 inducible gene) | ||
| phosphate | Cre12.g515750.t1.2 | Sodium-dependent phosphate transporter-related | ||
| Cre08.g379550.t1.2 | Sodium-dependent phosphate transporter, major facilitator superfamily | |||
| Cre12.g489400.t1.1 | PTB7 | Putative phosphate transporter, sodium/phosphate transporter | ||
| Cre02.g144650.t1.1 | PTB12 | Sodium/phosphate symporter | ||
| unspecified anions | Cre09.g404100.t1.1 | Cl- channel CLC-7 and related proteins (CLC superfamily) | ||
| Cre17.g729450.t1.1 | Cl- channel CLC-7 and related proteins (CLC superfamily) | |||
| Cre01.g037150.t1.2 | Voltage-gated chloride channel activity | |||
| sugars | Cre03.g206800.t1.2 | HXT1 | Hexose transporter | |
| P- and V-ATPases | Cre03.g176250.t1.1 | ATPvD1 | Vacuolar ATP synthase subunit D | |
| Cre10.g446550.t1.1 | ATPvF | Vacuolar ATP synthase subunit F | ||
| Cre01.g027800.t1.1 | ATPvH | Vacuolar ATP synthase subunit H | ||
| Mitochondrial electron transport / ATP synthesis | cytochrome c reductase | Cre01.g051900.t1.1 | RIP1 | Rieske iron-sulfur protein of mitochondrial ubiquinol-cytochrome c reductase (complex III) |
| Cre06.g262700.t1.2 | Ubiquinol cytochrome c reductase, subunit 7 | |||
| F1-ATPase | Cre02.g116750.t1.2 | F0F1-type ATP synthase, alpha subunit | ||
| Cre01.g018800.t1.1 | ATP6 | Mitochondrial F1F0 ATP synthase subunit 6 | ||
| Cre10.g420700.t1.1 | Mitochondrial F1F0-ATP synthase, subunit epsilon/ATP15 | |||
| Cre16.g680000.t1.1 | ATP5 | Mitochondrial ATP synthase subunit 5, OSCP subunit | ||
| NADH-DH | Cre10.g434450.t1.1 | NUOA9 | Putative NADH:ubiquinone oxidoreductase (Complex I) 39 kDa subunit | |
| Cre08.g378900.t1.1 | NUO3 | NADH:ubiquinone oxidoreductase ND3 subunit | ||
| Cre10.g450400.t1.1 | NUO5 | NADH:ubiquinone oxidoreductase (Complex I) 24 kD subunit | ||
| Lipid metabolism | 'exotics' (steroids, squalene etc) | Cre14.g615050.t1.1 | 3-oxo-5-alpha-steroid 4-dehydrogenase, Steroid reductase required for elongation of the VLCFAs (enoyl reductase) | |
| Cre12.g530550.t1.2 | KDG2 | Diacylglycerol kinase, sphingosine kinase | ||
| Cre02.g137850.t1.1 | NF* | |||
| FA desaturation | Cre17.g711150.t1.1 | Omega-6 fatty acid desaturase (delta-12 desaturase) | ||
| glyceral metabolism | Cre13.g577450.t1.2 | Glycerol-3-phosphate dehydrogenase | ||
| glycolipid synthesis | Cre13.g583600.t1.1 | DGD1 | Digalactosyldiacylglycerol synthase | |
| Cre16.g656400.t1.1 | SQD1 | UDP-sulfoquinovose synthase | ||
| lipid degradation | Cre06.g252801.t1.2 | CGI-141-related/lipase containing protein (TAG lipase) | ||
| Cre03.g164350.t1.2 | Lysophospholipase, putative drug exporter of the RND superfamily | |||
| phospholipid synthesis | Cre06.g281250.t1.1 | CFA1 | Cyclopropane fatty acid synthase | |
| Cre09.g398700.t1.1 | CFA2 | Cyclopropane fatty acid synthase | ||
| Cre11.g472700.t1.1 | NF* | |||
| Cre06.g262550.t1.1 | Zinc finger MYND domain containing protein 10 | |||
| Photosynthesis | Calvin-Benson cycle | Cre12.g511900.t1.1 | RPE1 | Ribulose phosphate-3-epimerase |
| Cre02.g120100.t1.1 | RBCS1 | Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit 1 | ||
| light reaction | Cre05.g243800.t1.1 | CPLD45 | Photosystem II Psb27 protein | |
| Cre10.g420350.t1.1 | PSAE | Photosystem I reaction center subunit IV | ||
| Cre01.g071450.t1.2 | NF* | |||
| Cre06.g291650.t1.1 | Ferredoxin | |||
| Cre05.g242400.t1.1 | No functional annotation | |||
| photorespiration | Cre09.g411900.t1.2 | SHMT3 | Serine hydroxymethyltransferase 3 | |
| Cre06.g295450.t1.1 | HPR1 | Hydroxypyruvate reductase | ||
| Major CHO metabolism | degradation | Cre09.g415600.t1.2 | Starch binding domain | |
| Cre11.g473500.t1.2 | NF* | |||
| Cre09.g415600.t1.2 | Starch binding domain | |||
| synthesis | Cre06.g289850.t1.2 | SBE1 | Starch Branching Enzyme | |
| Cre17.g721500.t1.1 | Granule-bound starch synthase I | |||
| misc | acid and other phosphatases | Cre13.g568600.t1.2 | Multiple inositol polyphosphate phosphatase-related, Acid phosphatase activity | |
| alcohol dehydrogenases | Cre13.g569350.t1.1 | Sterol dehydrogenase-related, Flavonol reductase/cinnamoyl-CoA reductase | ||
| cytochrome P450 | Cre07.g356250.t1.2 | Cytochrome P450 CYP4/CYP19/CYP26 subfamilies, beta-carotene 15,15'-monooxygenase | ||
| Cre07.g356250.t1.2 | Cytochrome P450 CYP4/CYP19/CYP26 subfamilies, beta-carotene 15,15'-monooxygenase | |||
| dynamin | Cre17.g724150.t1.1 | DRP3 | Dynamin-related GTPase | |
| GCN5-related N-acetyltransferase | Cre16.g657150.t1.2 | N-acetyltransferase activity (GNAT) family | ||
| gluco-, galacto- and mannosidases | Cre03.g171050.t1.2 | GHL1 | Glycosyl hydrolase | |
| misc2 | Cre14.g614100.t1.1 | GTR26 | Dolichyl-diphosphooligosaccharide-protein glycosyltransferase | |
| rhodanese | Cre07.g352550.t1.1 | RDP3 | Putative rhodanese domain phosphatase | |
| short chain dehydrogenase/reductase (SDR) | Cre07.g352450.t1.1 | Corticosteroid 11-beta-dehydrogenase and related short chain-type dehydrogenases, 3-hydroxybutyrate dehydrogenase | ||
| Cre12.g559350.t1.1 | 1-Acyl dihydroxyacetone phosphate reductase and related dehydrogenases | |||
| Cre03.g191850.t1.1 | Short chain dehydrogenase | |||
| UDP glucosyl and glucoronyl transferases | Cre11.g474450.t1.1 | NF* | ||
| Cre03.g205250.t1.2 | ELG4 | Exostosin-like glycosyltransferase | ||
| Cre16.g659500.t1.1 | Lactosylceramide 4-alpha-galactosyltransferase | |||
| Cre11.g483400.t1.2 | ELG10 | Exostosin-like glycosyltransferase | ||
| Tetrapyrrole synthesis | Glu-tRNA synthetase | Cre12.g510800.t1.1 | CHLI2 | Magnesium-chelatase subunit chlI |
| magnesium protoporphyrin IX methyltransferase | Cre12.g498550.t1.2 | Magnesium protoporphyrin IX S-adenosyl methionine O-methyl transferase (Magnesium-protoporphyrin IX methyltransferase) (PPMT) | ||
| unspecified | Cre12.g516350.t1.1 | COX10 | Cytochrome c oxidase assembly protein Cox10 | |
| urogen III methylase | Cre02.g133050.t1.2 | NF* | ||
-
*
Corresponding gene model was not found in v5.
SAK1 domain containing proteins in chlorophytes
| Number in alignment | Organism | Transcript/Protein IDaTranscript/Protein IDaTranscript/Protein ID* |
|---|---|---|
| 1 | Volvox carteri | Vocar20009235 |
| 2 | Volvox carteri | Vocar20002437 |
| 3 | Volvox carteri | Vocar20002672 |
| 4 | Volvox carteri | Vocar20004923 |
| 5 | Volvox carteri | Vocar20012349 |
| 6 | Volvox carteri | Vocar20005988 |
| 7 | Volvox carteri | Vocar20007158 |
| 8 | Volvox carteri | Vocar20007883 |
| 9 | Coccomyxa subellipsoidea | 57405 |
| 10 | Coccomyxa subellipsoidea | 59655 |
| 11 | Coccomyxa subellipsoidea | 57694 |
| 12 | Chlamydomonas reinhardtii | Cre16.g652650.t1.3 |
| 13 | Chlamydomonas reinhardtii | Cre06.g271000.t1.2 |
| 14 | Chlamydomonas reinhardtii | Cre06.g285800.t1.2 |
| 15 | Chlamydomonas reinhardtii | Cre06.g275600.t1.2 |
| 16 | Chlamydomonas reinhardtii | Cre06.g285750.t1.3 |
| 17 | Chlamydomonas reinhardtii | Cre06.g270950.t1.2 |
| 18 | Chlamydomonas reinhardtii | g9774.t1 |
| SAK1 | Chlamydomonas reinhardtii | KF985242 |
| 20 | Chlamydomonas reinhardtii | Cre03.g179150.t1.2 |
| 21 | Chlamydomonas reinhardtii | g3701.t1 |
| 22 | Chlamydomonas reinhardtii | Cre03.g179250.t1.2 |
| 23 | Chlamydomonas reinhardtii | Cre03.g179200.t1.2 |
| 24 | Chlamydomonas reinhardtii | Cre01.g004800.t1.2 |
| 25 | Chlamydomonas reinhardtii | Cre01.g048550.t1.3 |
| 26 | Chlorella variabilis | EFN51260 |
| 27 | Chlorella variabilis | EFN53496 |
| 28 | Chlorella variabilis | EFN55618 |
| 29 | Chlorella variabilis | EFN57652 |
| 30 | Chlorella variabilis | EFN55658 |
| 31 | Chlorella variabilis | EFN54262 |
| 32 | Chlorella variabilis | EFN54510 |
| 33 | Chlorella variabilis | EFN55806 |
| 34 | Chlorella variabilis | EFN53492 |
| 35 | Micromonas sp. RCC299 | ACO61347 |
| 36 | Micromonas pusilla CCMP1545 | EEH57791 |
| 37 | Micromonas sp. RCC299 | ACO65814 |
-
*
1–25, as defined on phytozome.net; 26–37, CrSAK1, genbank accession numbers.
Genes up-regulated during both 1O2 acclimation and dark to light transition
| Gene ID (v4) | Gene name | Annotation | RB (log2) | DL (log2) (Duanmu et al., 2013) |
|---|---|---|---|---|
| Cre02.g137700.t1.1* | 6.49 | 2.34 | ||
| Cre06.g281250.t1.1* | CFA1 | cyclopropane fatty acid synthase | 5.92 | 4.49 |
| Cre01.g033300.t1.1* | 5.72 | 3.62 | ||
| Cre13.g566850.t1.1* | SOUL2 | SOUL heme-binding protein | 5.53 | 2.25 |
| Cre13.g600650.t1.1* | 4.76 | 3.26 | ||
| Cre06.g263550.t1.1* | LCI7 | R53.5-related protein | 4.46 | 5.27 |
| Cre07.g342100.t1.1* | 4.43 | 1.84 | ||
| Cre09.g398700.t1.1* | CPLD27 | coclaurine N-methyltransferase | 4.05 | 1.36 |
| Cre12.g492650.t1.1* | FAS2 | fasciclin-like protein | 4.01 | 9.24 |
| Cre08.g381510.t1.1* | 3.94 | 3.27 | ||
| Cre10.g458450.t1.2* | GPX5 | glutathione peroxidase | 3.91 | 3.08 |
| Cre11.g474600.t1.1* | 3.90 | 1.99 | ||
| Cre13.g600700.t1.1* | 3.78 | 5.79 | ||
| Cre14.g613950.t1.1* | 3.65 | 2.68 | ||
| Cre06.g269300.t1.1* | 3.50 | 1.99 | ||
| Cre08.g380300.t1.2* | MSRA3 | peptide methionine sulfoxide reductase | 3.45 | 1.79 |
| Cre01.g031650.t1.2* | CGLD12 | protein with potential galactosyl transferase activity | 3.30 | 4.90 |
| Cre14.g629061.t1.1* | 3.25 | 1.88 | ||
| Cre13.g564900.t1.1* | 3.22 | 3.38 | ||
| Cre13.g586450.t1.1 | 3.21 | 3.50 | ||
| Cre02.g139500.t1.1* | 3.04 | 2.12 | ||
| Cre19.g756100.t1.1 | 3.04 | 6.53 | ||
| Cre01.g036000.t1.2 | 3.02 | 1.16 | ||
| Cre14.g618400.t1.1* | 2.97 | 2.16 | ||
| Cre17.g741300.t1.2* | 2.88 | 1.92 | ||
| Cre16.g648700.t1.2* | 2.79 | 2.35 | ||
| Cre17.g729950.t1.1 | 2.77 | 2.61 | ||
| Cre17.g721000.t1.1 | 2.70 | 2.12 | ||
| Cre06.g263500.t1.1* | 2.67 | 3.37 | ||
| Cre01.g016150.t1.1* | 2.65 | 2.92 | ||
| Cre08.g380000.t1.1* | 2.59 | 3.74 | ||
| Cre04.g224800.t1.1 | VAMP74 | R-SNARE protein, VAMP72-family | 2.58 | 3.34 |
| Cre03.g210150.t1.1 | 2.57 | 3.44 | ||
| Cre14.g615600.t1.1* | 2.53 | 2.40 | ||
| Cre06.g293100.t1.1 | Qc-SNARE SYP6-like protein | 2.48 | 4.90 | |
| Cre08.g368950.t1.1 | DHQS | 3-dehydroquinate synthase | 2.39 | 2.49 |
| Cre10.g424350.t1.2 | metalloprotease | 2.37 | 3.18 | |
| Cre12.g537225.t1.1 | 2.34 | 3.39 | ||
| Cre07.g336900.t1.2 | 2.32 | 2.31 | ||
| Cre16.g664050.t1.1 | 2.31 | 1.88 | ||
| Cre16.g677750.t1.1 | 2.04 | 2.22 | ||
| Cre12.g537227.t1.1 | 2.00 | 3.46 | ||
| Cre17.g737050.t1.1 | RabGAP/TBC protein | 1.99 | 2.32 | |
| Cre06.g297450.t1.1 | 1.93 | 1.46 | ||
| Cre06.g258600.t1.1* | 1.91 | 3.63 | ||
| Cre16.g663950.t1.1 | SC5D, C-5 sterol desaturase | 1.89 | 2.03 | |
| Cre13.g588150.t1.1 | 1.86 | 6.21 | ||
| Cre17.g722150.t1.1 | PKS3 | type III polyketide synthase | 1.85 | 1.61 |
| Cre16.g688550.t1.1 | GSTS1 | glutathione-S-transferase | 1.84 | 1.20 |
| Cre03.g207800.t1.1 | 1.84 | 7.09 | ||
| Cre10.g444550.t1.1* | SPP1A | signal peptide peptidase | 1.81 | 5.33 |
| Cre13.g602500.t1.2 | 1.76 | 1.59 | ||
| Cre03.g163400.t1.2* | 1.76 | 2.15 | ||
| Cre10.g450000.t1.1 | 1.74 | 2.18 | ||
| Cre01.g015500.t1.1 | 1.72 | 1.55 | ||
| Cre02.g105750.t1.2 | 1.71 | 3.23 | ||
| Cre01.g061750.t1.1 | SPT2 | serine palmitoyltransferase | 1.71 | 2.29 |
| Cre83.g796250.t1.1 | 1.68 | 1.59 | ||
| Cre16.g656150.t1.1 | 1.67 | 3.55 | ||
| Cre01.g002050.t1.2 | 1.66 | 3.15 | ||
| Cre12.g556750.t1.2 | Tic32-like 1 | Short-chain dehydrogenase, classical family, similar to PsTic32 | 1.66 | 3.15 |
| Cre12.g559100.t1.1 | 1.66 | 3.11 | ||
| Cre09.g411750.t1.2 | 1.61 | 1.96 | ||
| Cre11.g482650.t1.2 | 1.57 | 3.40 | ||
| Cre06.g310500.t1.1* | 1.57 | 6.23 | ||
| Cre09.g397900.t1.1 | transmembrane protein | 1.56 | 2.02 | |
| Cre04.g215600.t1.1 | 1.53 | 2.64 | ||
| Cre02.g093800.t1.1 | 1.51 | 4.99 | ||
| Cre02.g093750.t1.1 | NRX2 | Nucleoredoxin 2 | 1.50 | 6.26 |
| Cre01.g004350.t1.1 | 1.50 | 2.29 | ||
| Cre01.g034600.t1.1 | 1.50 | 2.22 | ||
| Cre11.g472600.t1.2 | 1.48 | 2.00 | ||
| Cre12.g500500.t1.2 | SMT1 | sterol-C24-methyltransferase | 1.46 | 3.05 |
| Cre13.g577950.t1.1 | VPS6 | subunit of the ESCRT-III complex | 1.45 | 2.36 |
| Cre02.g118200.t1.1 | 1.44 | 2.79 | ||
| Cre01.g012500.t1.1 | PRA1 | prenylated rab acceptor family protein | 1.43 | 2.46 |
| Cre12.g521600.t1.2 | 1.42 | 2.89 | ||
| Cre03.g179100.t1.1 | ubiquitin fusion degradation protein | 1.41 | 3.38 | |
| Cre09.g413150.t1.2 | 1.39 | 4.31 | ||
| Cre13.g572200.t1.1 | tyrosine/tryptophan transporter protein | 1.39 | 2.57 | |
| Cre03.g185850.t1.2 | PfkB-type carbohydrate kinase | 1.37 | 3.05 | |
| Cre18.g743600.t1.1 | 1.37 | 1.65 | ||
| Cre02.g076800.t1.1 | sterol reductase | 1.36 | 2.41 | |
| Cre06.g256750.t1.1 | FAT1 | acyl carrier protein thioesterase | 1.35 | 1.67 |
| Cre17.g729450.t1.1 | 1.34 | 1.90 | ||
| Cre11.g471550.t1.1 | 1.34 | 3.29 | ||
| Cre09.g395750.t1.2 | 1.33 | 2.87 | ||
| Cre14.g617100.t1.1 | 1.33 | 3.33 | ||
| Cre16.g691500.t1.1 | Sec14p-like lipid-binding protein | 1.33 | 2.28 | |
| Cre02.g079550.t1.1 | DRP2 | Dynamin-related GTPase | 1.32 | 2.34 |
| Cre02.g079300.t1.1 | VPS4 | AAA-ATPase of VPS4/SKD1 family | 1.32 | 1.96 |
| Cre05.g231700.t1.2 | 1.31 | 2.40 | ||
| Cre02.g132300.t1.2 | DNJ12 | DnaJ-like protein | 1.30 | 2.24 |
| Cre69.g794101.t1.1 | 1.30 | 2.65 | ||
| Cre13.g565600.t1.2 | 1.29 | 3.42 | ||
| Cre13.g593700.t1.1 | monooxygenase, DBH-like | 1.29 | 1.81 | |
| Cre12.g498000.t1.2 | 1.28 | 3.88 | ||
| Cre06.g292900.t1.2 | 1.28 | 2.16 | ||
| Cre08.g372100.t1.1 | HSP70A | Heat shock protein 7A | 1.27 | 2.28 |
| Cre01.g039350.t1.1 | NCR2 | NADPH-cytochrome P45 reductase | 1.26 | 2.19 |
| Cre03.g211100.t1.1 | 1.26 | 2.11 | ||
| Cre17.g731800.t1.1 | 1.25 | 1.78 | ||
| Cre17.g730650.t1.1 | 1.25 | 2.28 | ||
| Cre02.g123000.t1.2 | 1.24 | 1.42 | ||
| Cre05.g247700.t1.2 | 1.24 | 2.71 | ||
| Cre08.g360800.t1.2 | haloacid dehalogenase-like hydrolase | 1.23 | 4.39 | |
| Cre07.g350750.t1.1 | PTOX1 | alternative oxidase | 1.22 | 3.32 |
| Cre17.g703750.t1.1 | 1.20 | 2.21 | ||
| Cre06.g306041.t1.1 | 1.20 | 2.90 | ||
| Cre02.g116650.t1.1 | 1.20 | 2.83 | ||
| Cre08.g379400.t1.2 | 1.18 | 3.04 | ||
| Cre16.g677000.t1.1 | HSP70E | Heat shock protein 7E | 1.18 | 2.50 |
| Cre06.g283900.t1.1 | 1.18 | 5.24 | ||
| Cre14.g626750.t1.1 | 1.17 | 4.12 | ||
| Cre01.g010700.t1.1 | 1.16 | 2.10 | ||
| Cre01.g002000.t1.2 | predicted proteim | 1.15 | 1.68 | |
| Cre04.g213150.t1.1 | 1.15 | 2.78 | ||
| Cre16.g694250.t1.1 | 1.15 | 2.92 | ||
| Cre05.g246400.t1.1 | 1.15 | 2.74 | ||
| Cre02.g128450.t1.1 | 1.13 | 2.82 | ||
| Cre03.g180250.t1.1 | Myo-inositol-1-phosphate synthase | 1.13 | 2.05 | |
| Cre03.g186150.t1.1 | 1.13 | 1.78 | ||
| Cre02.g137800.t1.1 | 1.13 | 2.00 | ||
| Cre11.g471500.t1.1 | MFT10 | predicted protein | 1.11 | 1.40 |
| Cre10.g435200.t1.1 | 1.10 | 2.13 | ||
| Cre13.g593850.t1.2 | 1.10 | 3.91 | ||
| Cre19.g754000.t1.2 | 1.10 | 2.33 | ||
| Cre13.g593869.t1.1 | 1.10 | 3.90 | ||
| Cre08.g377300.t1.2 | 1.09 | 3.27 | ||
| Cre04.g225050.t1.2 | predicted protein | 1.09 | 3.55 | |
| Cre07.g330300.t1.1 | 1.08 | 2.22 | ||
| Cre12.g500450.t1.2 | 1.08 | 3.00 | ||
| Cre06.g262000.t1.1 | 1.08 | 1.87 | ||
| Cre10.g441550.t1.2 | MAM3B | predicted protein | 1.07 | 1.54 |
| Cre06.g249800.t1.1 | unknown conserved protein | 1.07 | 2.08 | |
| Cre01.g038250.t1.1 | SDC1 | serine decarboxylase | 1.06 | 1.92 |
| Cre44.g788200.t1.1 | 1.06 | 2.13 | ||
| Cre08.g359200.t1.2 | 1.03 | 2.69 | ||
| Cre05.g245950.t1.1 | DRP1 | Dynamin-related GTPase | 1.03 | 2.15 |
| Cre05.g234100.t1.1 | CYP745A1 | cytochrome P45 | 1.01 | 2.61 |
| Cre07.g328700.t1.2 | 1.01 | 1.56 | ||
| Cre10.g440250.t1.2 | 1.01 | 2.14 | ||
| Cre17.g725200.t1.1 | MDR-like ABC transporter | 1.01 | 3.30 | |
| Cre82.g796100.t1.1 | 1.01 | 2.49 |
-
*
Genes defined as SAK1-dependent in Table 4.
Primers used for qRT-PCR analyses
| v4 ID | v5 ID | Gene name | Forward | Reverse |
|---|---|---|---|---|
| Cre01.g007300.t1.1 | Cre01.g007300.t1.2 | AGCATGTGCGTGTGGAGTAG | CCTTACCATAGGCCTGACCA | |
| au5.g10700_t1a | Cre03.g177600.t1.3 | CTGGACATGTCGGCTATGAA | GCTCATGTCGTACTCCAGCA | |
| au5.g13389_t1* | Cre06.g299700.t1 | SOUL1† | TGCGTATGGGTGTCCACTAA | TGGGGATCTTCTTCATGTCC |
| Cre06.g263550.t1.1 | Cre06.g263550.t1.2 | LCI7 | TTTGGTTGCGTTGCATGTAT | TCAACGCGGTGTCAAACTTA |
| Cre06.g281250.t1.1 | Cre06.g281250.t1.2 | CFA1 | CCTACAACGACAACGACGTG | GGAAGTTCCAGGATGACCAG |
| Cre06.g298750.t1.1 | Cre06.g298750.t1.2 | AOT4 | CCGTGTGCACAGATTCAAAG | CACACAGCGCCTCCTACATA |
| Cre08.g358200.t1.2 | Cre08.g358200.t2.1 | TGTGGCATCAAGGTGTGTTGT | AACCCCACACCCCTCTCTTT | |
| Cre09.g398700.t1.1 | Cre09.g398700.t1.2 | CFA2 | CGACCTGCTGCTCTACTTCC | GTGTAGGCGGTGGTCAAGAT |
| Cre10.g458450.t1.2 | Cre10.g458450.t1.3 | GPX5 | AACCAATCGCCTAACACCTG | CACTTGCTAGCCACGTTCAC |
| Cre12.g503950.t1.1 | Cre12.g503950.t1.2 | GGAGGGAGTACCACGAGACA | GATTGCTGTAAGGCCGGATA | |
| Cre13.g564900.t1.1 | Cre13.g564900.t1.2 | MRP3 | TCATGACGTACATCTCGATTCTCA | AGGGAATGTAGTAGCGCTGAATG |
| au5.g4402_t1* | Cre13.g566800.t1.2 | TGCTTGGAAGACCCACTTTT | GAGCTGGAGTTGCAGTTGTG | |
| Cre13.g566850.t1.1 | Cre13.g566850.t1.2 | SOUL2 | CCCTCCCCTCCTTCAGACTA | CGTACCTGAGGCGCATATTT |
| Cre14.g613950.t1.1 | Cre14.g613950.t2.1 | CGCCCAACCCCATGATC | CCGCAACGTACCGTGATG | |
| Cre16.g683400.t1.1 | Cre16.g683400.t1.2 | CCTGAACAAACACACGATGG | GAACGCCGTCAAATCATCTT | |
| Cre16.g688550.t1.1 | Cre16.g688550.t1.2 | GST1 | AGTGCGGAGGAAGTCGTAAA | GTAAAAGACGTGCGTGCAAA |
| g6364.t1 | CβLP(RCK1) | GAGTCCAACTACGGCTACGC | GGTGTTCAGGTCCCACAGAC | |
| Cre14.g623650.t1.1 | Cre14.g623650.t1 | GACAACGCGGCCTACAAGA | CCGAGCTGGCGGTGTTAA | |
| au5.g2281_t1* | g16723.t1 | MKS1 | GCTTGAGCGCGAGACGAA | CGCTGAAAGCATTGCAGAAG |
| Cre08.g380300.t1.2 | Cre08.g380300.t1.2 | ACCACCAGCAGTACCTGTCC | CGCTCCAATAAAGCCTTCAG | |
| au5.g7871_t1‡ | (Cre17.g741300.t1.2)‡ | SAK1(5'UTR) | CAAGTGCTCATGAGAGGCCTTA | TACGTCATCCAGTTCCACATCC |
| au5.g7871_t1‡ | (Cre17.g741300.t1.2)‡ | SAK1(3'UTR) | TCAAGCGTGTGGGTAAGAGCTA | ACGCTATCTCCGTCCTAATCCA |
| Cre08.g365900.t1.1 | Cre08.g365900.t1.2 | LHCSR1 | CACACAATTCTGCCAACAGC | ATCTGCTTCACGGTTTGGTC |
| Cre04.g220850.t1.1 | Cre04.g220850.t1.2 | TAATGGTATGGATGCGGTCA | ACTGCCAGTTATGGGTCCTG | |
| Cre09.g395750.t1.2 | Cre09.g395750.t1.3 | ACCGTCCGTGAACCTTACTG | CGCAAACACGTCTCAAAGAA |
-
*
Was originally mapped and identified as augustus version 5 models within Chlamydomonas genome v4.
-
†
SOUL1 was given the name in v4 but not v5.
-
‡
Primers were designed against experimentally obtained cDNA (Genbank accession KF985242) and differs from v5. Closest gene model is au5.g7871_t1.
Additional files
-
Supplementary file 1
Genes that display significant differential expression by pair-wise comparisons.
- https://doi.org/10.7554/eLife.02286.023





