AFF4 binding to Tat-P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV promoter
Figures
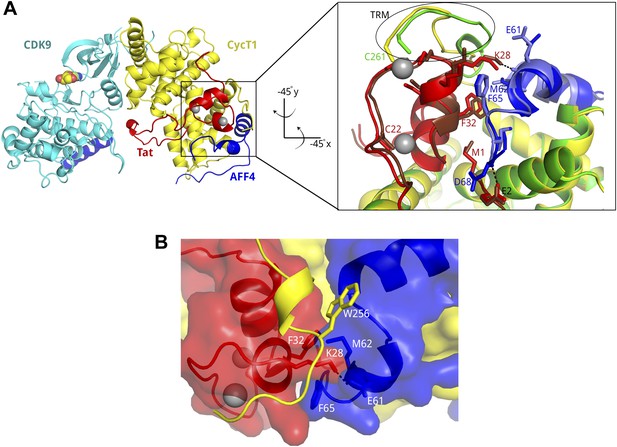
Tat and AFF4 bind adjacent to each other on the CycT1 surface.
(A) Tat-AFF4-P-TEFb ribbon diagram (left) showing interactions between Tat (red) and AFF4 (blue) bound to the CycT1 (yellow) subunit of P-TEFb. AFF4 helix 0 is bound to the CDK9 (cyan) subunit, and adenosine (spheres) is modeled in the CDK9 ATP binding pocket. The close-up view (right) obtained by horizontal and vertical 45° rotations of the left hand figure shows similar Tat–AFF4 (red/dark red-blue/light blue) interactions in independent complexes. CycT1 (yellow/green) TRM residues adopt different structures in different crystal environments. The upper of the two Zn2+ ions (gray spheres) anchors the CycT1 TRM. (B) Surface representation of the binding pocket for Tat K28 in the Tat–AFF4 (red-blue) interface. The CycT1 TRM (yellow ribbon) with the fewest crystal contacts is shown. The TRM interacts with a hybrid interface including AFF4 and Tat.
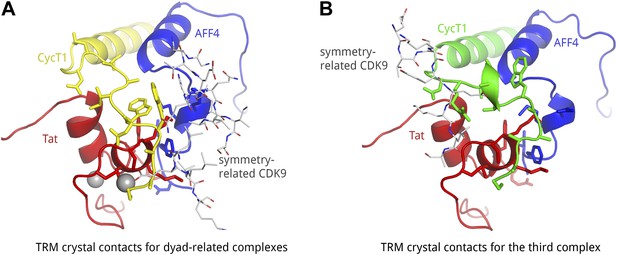
Crystal contacts of CycT1 TRM for two representative complexes in the a.u..
(A) In the two dyad-related complexes, CDK9 (gray sticks) molecules from adjacent complexes make contacts with the C-terminal end of the CycT1 (yellow) TRM, as well as AFF4 (blue). (B) In the third complex, the neighboring CDK9 (gray sticks) interacts with the N-terminus of the CycT1 (green) TRM. The C-terminal residues of the TRM are exposed to solvent.
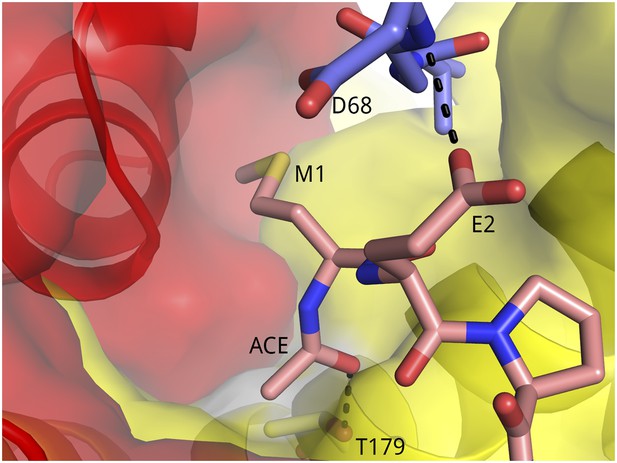
Surface representation of the binding pocket for Tat M1 and the N-acetyl group.
The methionine side chain binds in a pocket formed by Tat (red) and CycT1 (yellow).
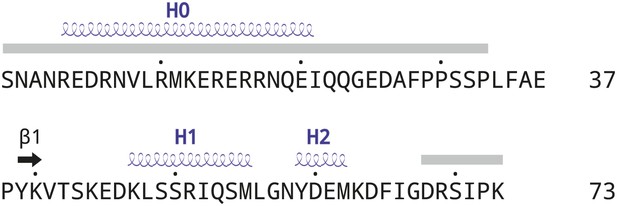
Schematic drawing of AFF4 secondary structures.
Disordered regions (gray rectangles), α helices (blue springs) and the short β strand (black arrow) are indicated. Helix H0 in AFF4 is only observed in two out of three molecules in the a.u.
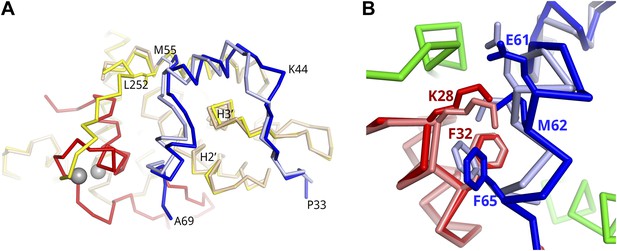
Structural shifts in the subunits of the Tat-AFF4-P-TEFb complex.
(A) Superposition of the AFF4-P-TEFb complex (PDB ID 4IMY, pastel colors) and Tat-AFF4-P-TEFb (red, blue, yellow) on the CycT1 subunit shows coupled shifts of the two AFF4 helices. AFF4 helices 1 and 2 shift away from Tat, thereby avoiding close contacts between Tat and helix 2. (B) Superposition of AFF4-P-TEFb (PDB ID 4IMY, AFF4 light blue), Tat-P-TEFb (PDB ID 3MI9, Tat pastel-red), and Tat-AFF4-P-TEFb (Tat red, AFF4 blue, CycT1 green) on the CycT1 subunit. CycT1 of AFF4-P-TEFb and Tat-P-TEFb is omitted to emphasize the changes in AFF4. The AFF4 backbone shifts 1–2 Å in the presence of Tat, while the Tat conformation displays only small changes associated with AFF4 binding. Side chains undergo only small conformational changes.
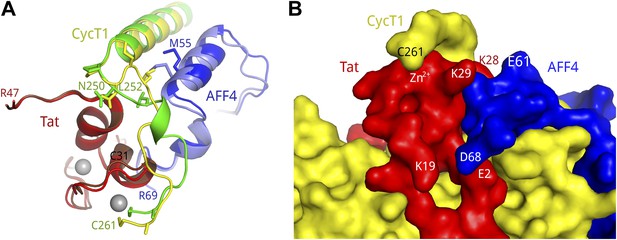
CycT1 TRM interacts with Tat and AFF4.
(A) Ribbon diagram of two distinct TRM conformations observed in the Tat-AFF4-P-TEFb crystal structure (red, blue, yellow/dark red, light blue, green). Zn2+ ions are shown as gray spheres. (B) Surface representation of Tat-AFF4-CycT1 interactions. The three subunits intertwine, thereby stabilizing the TRM conformation in the hybrid interface.
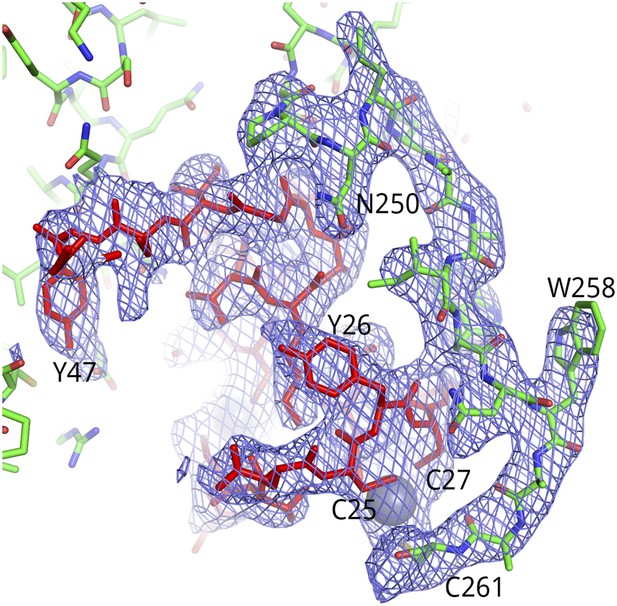
Representative electron density for the Tat-AFF4-P-TEFb complex.
2Fo-Fc map (1.0 σ) for Tat (red) and CycT1 TRM (green) is shown for a dyad-related complex. Residues of the CycT1 TRM were omitted from the model used for molecular replacement and subsequently built into the omit electron density.
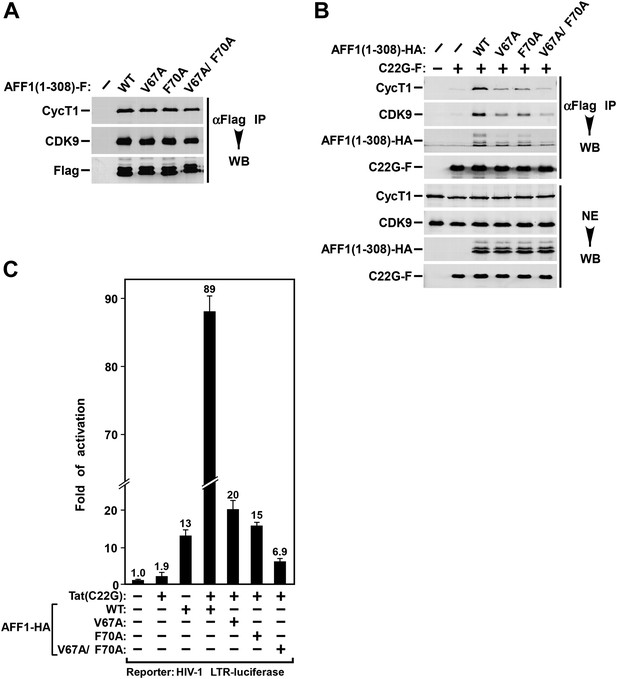
AFF1 Tat interaction mutants reduce Tat binding and activation of HIV LTR by AFF1.
(A) Nuclear extracts (NE) were prepared from HeLa cells expressing the truncated Flag-tagged AFF1 protein (residues 1–308). Anti-Flag immunoprecipitates (IP) from the NE were examined by Western blotting (WB) for the indicated proteins. (B) Nuclear extracts were prepared from HeLa cells co-expressing Flag-tagged Tat(C22G) and haemagglutinin (HA)-tagged truncated AFF1. Anti-Flag IPs were analyzed as in A. (C) HeLa-based NH1 cells containing the intergrated HIV-1 LTR-luciferase reporter gene were transfected with the Tat(C22G)- and/or AFF1-expressing construct as labeled. Luciferase activities were measured in cell extracts, with the level of activity detected in cells transfected with an empty vector (−) set to 1. The error bars represent mean ± SD from three independent measurements.
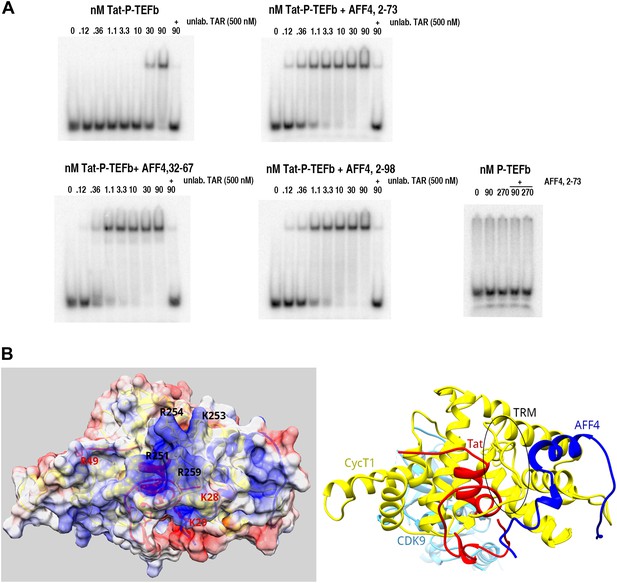
SECs stimulate TAR recognition.
(A) Electrophoretic mobility shift assays with 32P-labeled TAR and increasing concentrations of Tat-P-TEFb, or Tat-P-TEFb + AFF432–67, Tat-P-TEFb + AFF42–73, Tat-P-TEFb + AFF42–98. Control assays (bottom right) with P-TEFb and AFF42–73 showed no shifts for TAR. Half of TAR was shifted with 35–40 nM Tat-P-TEFb complex. In the presence of excess AFF4 fragments 32–67, 2–73, or 2–98, 50% of TAR was shifted by 1.1 nM Tat-AFF4-P-TEFb complex. (B) Calculated electrostatic surface potential of Tat-AFF4-CycT1 centered on the CycT1 TRM. The ribbon diagram (right) is in the same orientation as the surface representation (left). This orientation converts into the orientation in Figure 1A by consecutive rotations around y (70°) and z (−35°). CDK9 was omitted from the surface figure (left) to focus on the TAR interaction region. Solvent-exposed CycT1 residues K253, R254, N257, W258, R259, and Tat R49, which have no side-chain electron density, were modeled in the most common orientation. The electrostatic potential, calculated using APBS (Baker et al., 2001) was applied to color the solvent excluded surface of Tat-AFF4-CycT1 in Chimera (Pettersen et al., 2004) from −5 kbTe−1 (red) to +5 kbTe−1 (blue). CycT1 residues were labeled in black, Tat residues in red. The TRM region forms a positively charged patch on the SEC surface close to the disordered Tat ARM, which follows Tat R49.
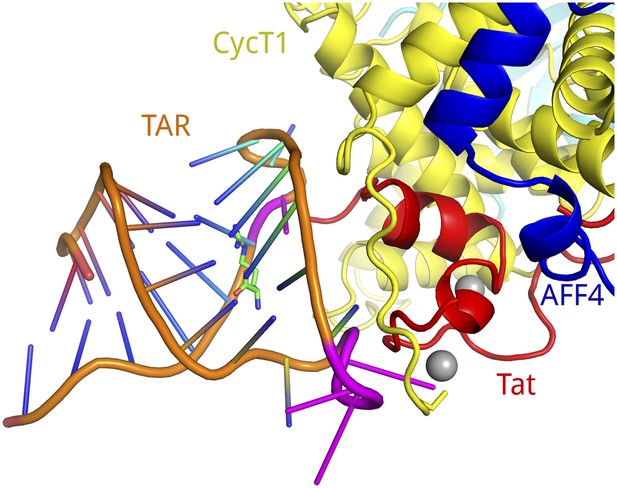
Model of TAR binding to SEC.
The positively charged CycT1 TRM is positioned close to the predicted location of the Tat ARM, which binds to bases in the TAR bulge (U23–U25) (Weeks and Crothers, 1991). Considering that the CycT1 TRM is interacting with the TAR loop region (C30–A35) (Richter et al., 2002), we manually placed the solution structure of TAR-arginimide (PDB ID 1ARJ) onto the Tat-AFF4-P-TEFb model so that the TAR bulge neighbors the Tat ARM region and the TAR loop contacts the CycT1 TRM. The dimensions of the components match well. The color scheme for the SEC is the same as in previous figures. The TAR phosphate backbone is shown in orange with bases in blue. TAR bases important for binding to Tat or CycT1 are drawn in magenta.
Tables
X-ray data collection and refinement statistics for P-TEFb-Tat-AFF4
| Data collection | |
| Space group | P6522 |
| Cell dimensions: a, b, c | 184.91, 184.91, 360.40 |
| Resolution (Å)* | 50.0–3.0 (3.05–3.0) |
| Unique reflections* | 73,424 (3589) |
| I/σ(I)* | 12.8 (0.9) |
| Rmerge (%)* | 22.2 (>100) |
| Rmerge (%)*, I/sigI≥3 | 8.4 (18.9) |
| Rpim (%)† | 7.6 (87.9) |
| CC1/2 high resolution shell | 0.553 |
| Completeness (%)* | 100.0 (100.0) |
| Redundancy* | 24.2 (23.8) |
| Temperature (K) | 100 |
| Mosaicity (°) | 0.23–0.39 |
| Refinement | |
| Resolution (Å) | 49.0–3.0 |
| No. reflections | 73,297 |
| Rwork/Rfree* | 0.206/0.232 (0.316/0.335) |
| No. atoms/B-factors (Å2) | |
| CDK9, molecule 1, 2, 3 | 2560 (75.4), 2521 (90.9), 2572 (88.5) |
| Cyclin T1, molecule 1, 2, 3 | 2061 (79.4), 2053 (85.8), 2058 (97.8) |
| AFF434-66, molecule 1, 2, 3 | 438 (85.0), 268 (115.7), 422 (92.3) |
| Tat | 390 (79.1), 384 (78.0), 390 (102.7) |
| Water | 37 (58.7) |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.0035 |
| Bond angles (°) | 0.811 |
| Ramachandran plot‡ | |
| Favored (%) | 96.0 |
| Allowed (%) | 3.36 |
| Disallowed (%) | 0.66 |
-
*
Values in parentheses are for the highest resolution shell.
-
†
Rp.i.m. is the precision-indicating merging R factor, which is related to the traditional Rsym but provides a better estimate of data quality (Weiss and Hilgenfeld, 1997; Weiss et al., 1998).
-
‡
Values from MOLPROBITY (Chen et al., 2009).





