Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation
Figures
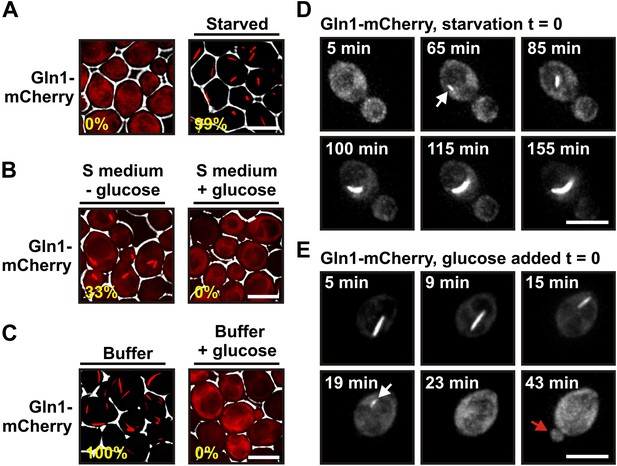
Gln1 assembles into filaments in energy-depleted yeast cells.
(A) Yeast cells expressing mCherry-tagged Gln1 from the endogenous promoter were washed twice with water and resuspended in synthetic media (left, control) or citrate buffer of pH 6 (right, ‘starved’). White lines are the cell boundaries. The scale bar is 5 μm. The numbers in yellow give the percentage of cells with fluorescent foci. At least 200 cells were counted. (B) Log phase yeast cells expressing mCherry-tagged Gln1 were washed twice with water and resuspended in synthetic media without (left) or with (right) 2% glucose. Images were taken 4 hr after onset of glucose starvation. (C) Log phase cells expressing mCherry-tagged Gln1 were washed twice with water and resuspended in a phosphate–citrate buffer of pH 6 without (left) or with (right) 2% glucose. Images were taken 4 hr after onset of starvation. (D) Cells expressing Gln1-mCherry were washed twice with water and resuspended in a phosphate–citrate buffer of pH 6 to induce starvation (time point 0). Filament formation was followed by time-lapse microscopy. Individual time points are indicated in minutes. The white arrow designates an emerging filament. The scale bar is 5 μm. Also see the corresponding Video 1. (E) Same as (D) except that filament dissolution was investigated by re-adding glucose to cells that had been starved for 4 hr. The white arrow points to a small filament. The red arrow designates the emerging bud. Also see the corresponding Video 3.
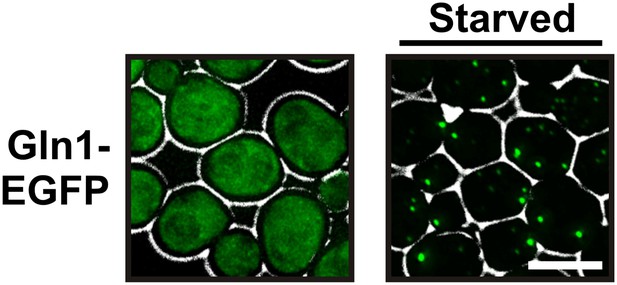
GFP-tagged Gln1 predominantly forms punctate structures.
Yeast cells expressing GFP-tagged Gln1 from the endogenous promoter were washed twice with water and resuspended in synthetic media (left, control) or buffer of pH 6 (right, ‘starved’). White lines are the cell boundaries. The scale bar is 5 μm.
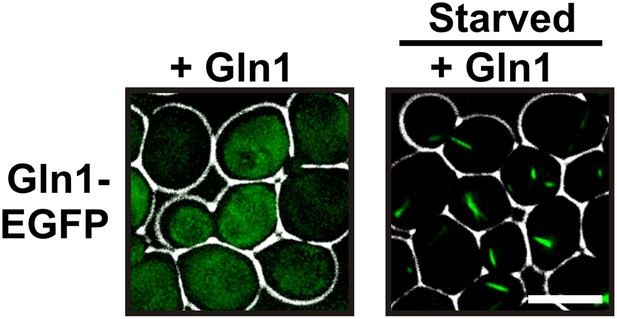
Co-expression of untagged Gln1 transforms the localization pattern from punctate to filamentous.
Yeast cells expressing GFP-tagged Gln1 from the endogenous promoter were washed twice with water and resuspended in synthetic media (left, control) or buffer of pH 6 (right, ‘starved’). The cells co-expressed untagged Gln1 from a plasmid. White lines are the cell boundaries. The scale bar is 5 μm.
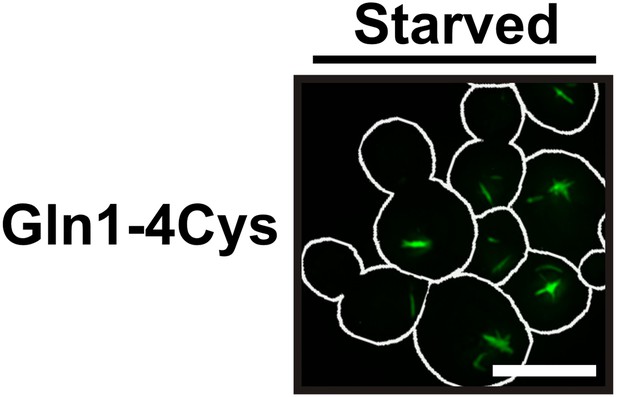
Filamentation is not caused by the tag.
Yeast cells expressing tetracystein-tagged Gln1 were incubated over night with FIAsH-EDT2 to label Gln1. The cells were washed twice with water and resuspended in a phosphate–citrate buffer to induce starvation (pH 6). Images were taken 4 hr after onset of starvation. White lines denote the cell boundaries. The scale bar is 5 μm.
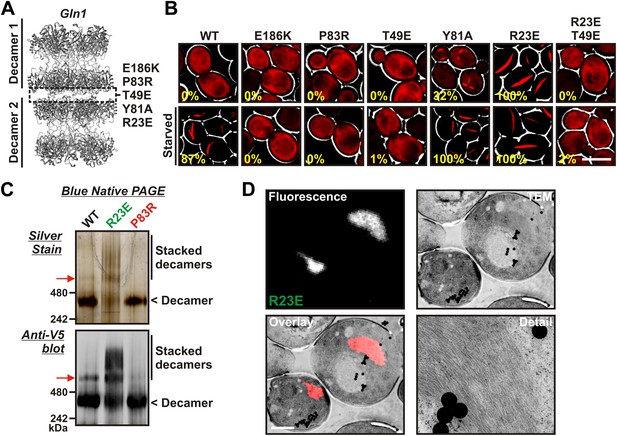
Gln1 assembles by a back-to-back stacking mechanism.
(A) The crystal structure of Gln1 as reported by He at al. (2009). The putative assembly interface and mutations introduced in this study are indicated. (B) Chromosomally encoded Gln1 was replaced with mCherry-tagged wild-type or variant Gln1 expressed from a plasmid. The strains were washed twice with water and resuspended in synthetic media (top, control) or buffer (bottom, ‘starved’). The numbers in yellow give the percentage of cells with fluorescent foci. At least 200 cells were counted. The scale bar is 5 μm. (C) Wild type or variant 6xHis-Gln1-V5 was purified from yeast and analyzed by blue native PAGE. Proteins were detected by silver staining (top) or immunoblotting with an antibody that recognized a C-terminal V5 tag (bottom). The red arrow denotes a band that corresponds to two stacked decamers. The calculated size of a Gln1 decamer is 420 KDa. (D) Correlative light electron microscopy (CLEM) was performed on yeast cells expressing the R23E variant of Gln1 as mCherry fusion. The black dots are fluorescent beads, which were introduced to facilitate the alignment of the fluorescence and TEM images. The scale bar is 500 μm.
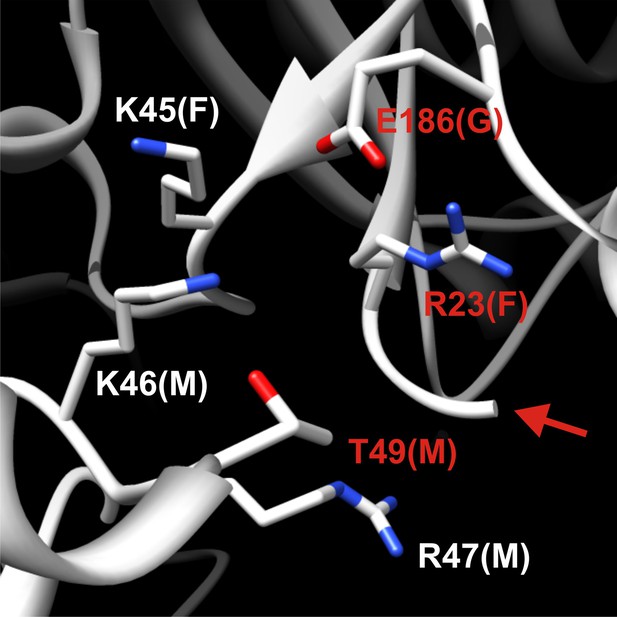
Detailed structural view of the decamer–decamer interface.
All critical residues are shown within a range of 10 Å. Residues mutated in this study are highlighted in red. The subunit identifier is given in brackets. The red arrow denotes the disordered N terminus. The mutations introduced were E186K, R23E, and T49E.
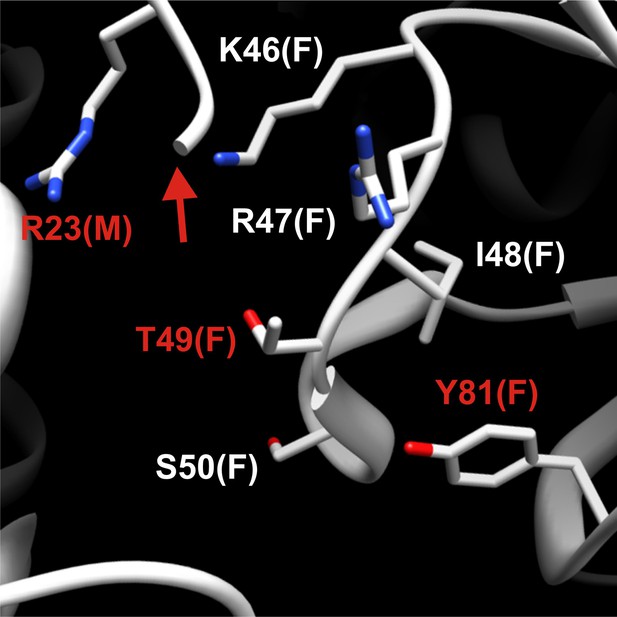
Detailed structural view of the decamer–decamer interface.
All critical residues are shown within a range of 10 Å. Residues mutated in this study are highlighted in red. The subunit identifier is given in brackets. The red arrow denotes the disordered N terminus. The mutations introduced were R23E, T49E, and Y81A.
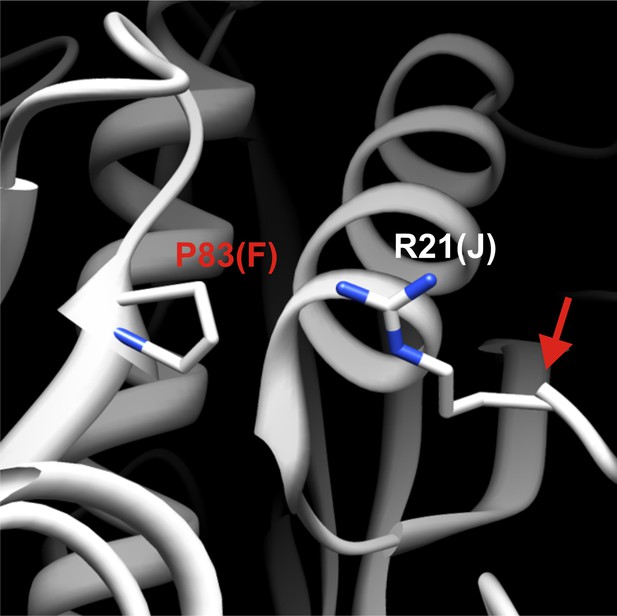
Detailed structural view of the decamer–decamer interface.
All critical residues are shown within a range of 10 Å. Residues mutated in this study are highlighted in red. The subunit identifier is given in brackets. The red arrow denotes the disordered N terminus. The mutation introduced was P83R.
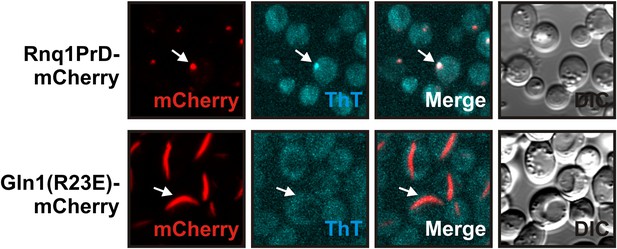
Yeast cells expressing mCherry-tagged R23E Gln1 were subjected to staining with Thioflavin T.
Only the control filaments (formed by the yeast prion Rnq1) were stainable by ThT, suggesting that the formation of cross-β structure is not required for Gln1 filamentation.
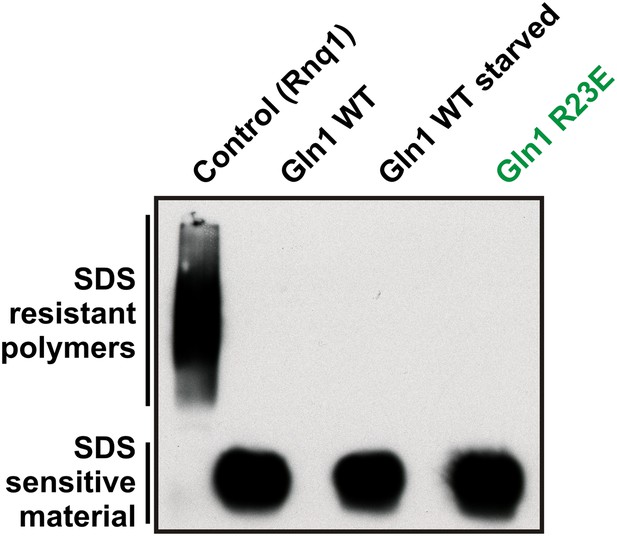
Lysates from yeast cells expressing wild-type or R23E Gln1 were subjected to semi-denaturing detergent-agarose gel electrophoresis (SDD-AGE).
Only the control filaments (formed by the yeast prion Rnq1) were resistant to SDS, indicating that Gln1 filamentation does not involve the formation of cross-β structure.
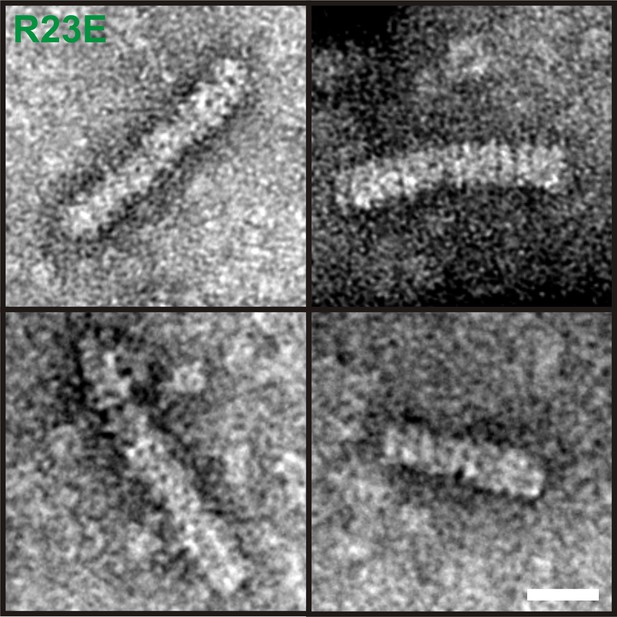
6xHis-Gln1(R23E)-mCherry was affinity purified from yeast, subjected to negative staining and investigated by electron microscopy.
Note the presence of filaments with a diameter of ∼120 Å and a repeating unit of ∼100 Å in size. These proportions are consistent with the reported dimensions of Gln1 decamers (He et al., 2009). The scale bar is 20 nm.
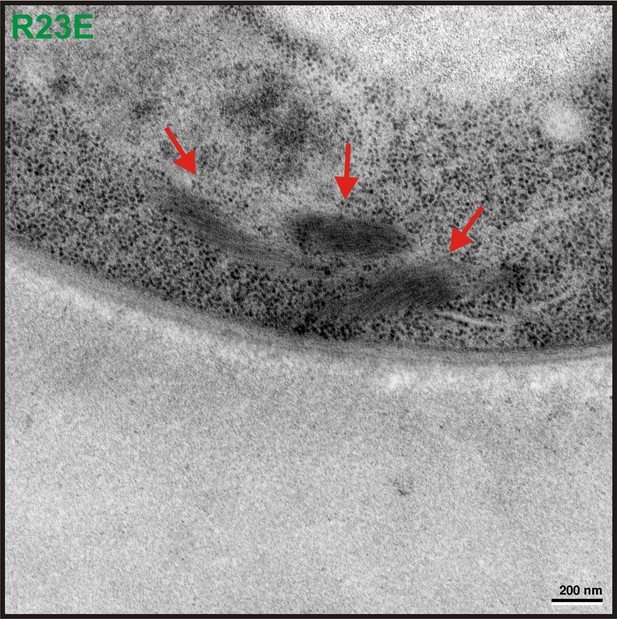
Transmission electron microscopy (TEM) was performed on yeast cells expressing untagged Gln1(R23E).
The red arrow points to filamentous structures in the cytoplasm. Similar structures were absent from control cells expressing untagged Gln1. The scale bar is 200 nm.
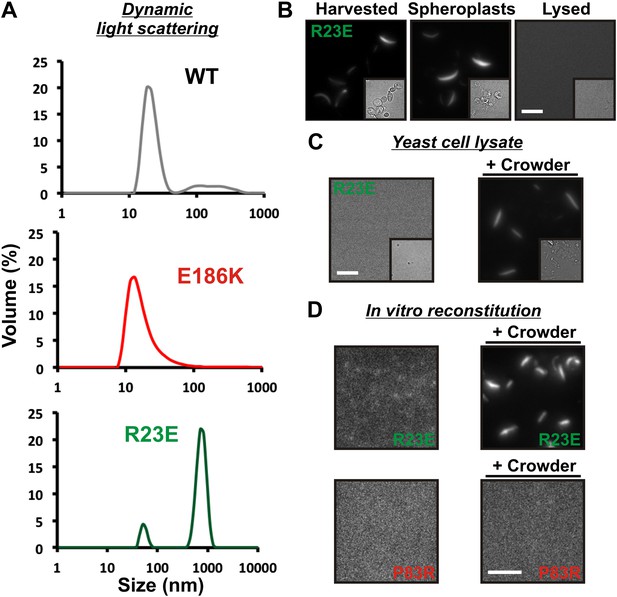
Self-assembly into filaments is driven by macromolecular crowding.
(A) Equal amounts of 6xHis-tagged wild-type and variant Gln1 purified from bacteria were subjected to dynamic light scattering. Shown is the volume distribution that was derived from the intensity distribution. Note the different scales of the x axes. (B) Yeast cells expressing Gln1(R23E)-mCherry were spheroplasted and lysed. Images were acquired from harvested, spheroplasted, and lysed cells. Images were taken at the same intensity settings. The inset is the corresponding DIC image. The scale bar is 5 μm. (C) Cells were treated as in (A) except that the lysis buffer contained Ficoll 70 at a concentration of 200 mg/ml. (D) Gln1-mCherry was purified from yeast and incubated in a phosphate-citrate buffer of pH 7 with or without a crowding agent for 1 hr. Samples were analyzed by fluorescence microscopy and images were taken at the same intensity settings. The scale bar is 3 μm.
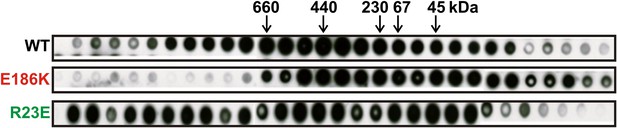
Gel filtration of wild type and variant 6xHis-tagged Gln1 purified from bacteria.
Fractions were applied onto a nitrocellulose filter by using a dot blot apparatus. Molecular weight markers were thyroglobulin (660 kDa), ferritin (440 kDa), catalase (230kD), bovine serum albumin (67 kDa), and ovalbumin (43 kDa).
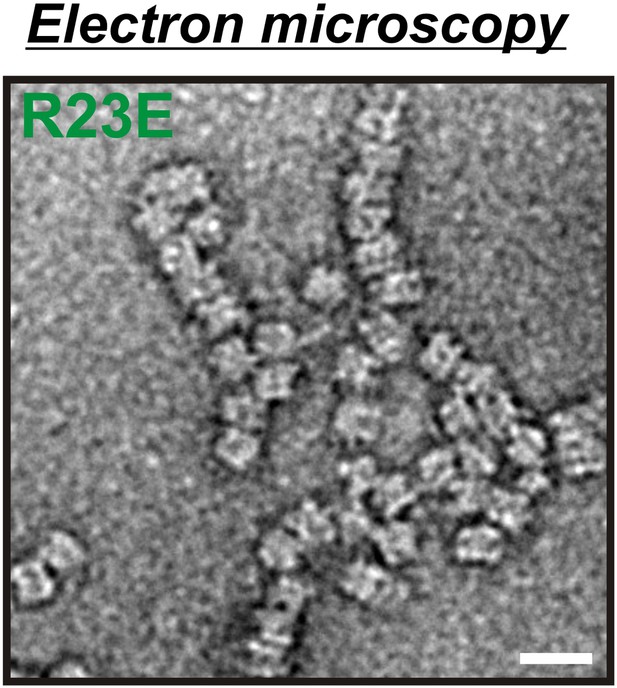
Electron microscopy of 6xHis-tagged R23E Gln1 purified from bacteria.
Note the formation of chains, which indicate enzyme stacking by a back-to-back mechanism. The scale bar is 20 nm.
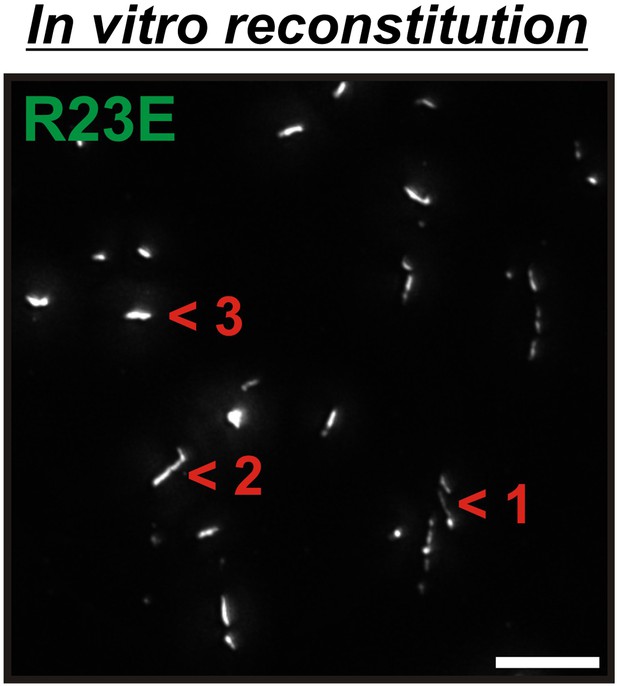
6xHis-Gln1-mCherry was affinity purified from yeast and assembled in the presence of a crowder.
The sample was investigated by fluorescence microscopy. Acquired images were deconvolved to increase the signal to noise ratio. The shown image is a maximum intensity projection of 20 individual images that were acquired with 200 nm sectioning. Note the presence of filamentous structures of varying thickness (numbers 1, 2 and 3), indicating the formation of higher order fibrils from laterally aligning filaments.
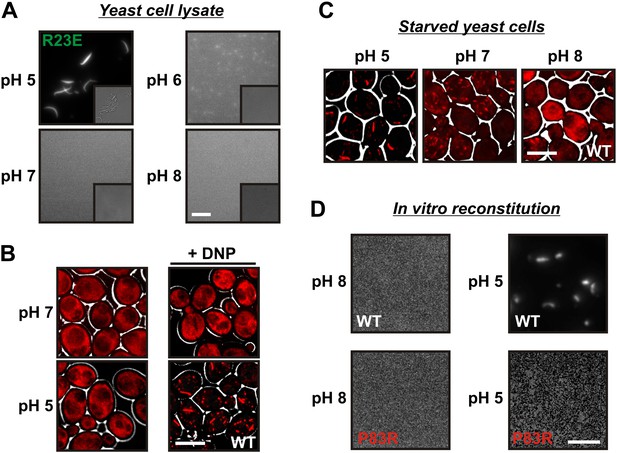
A drop in intracellular pH triggers filament formation.
(A) Yeast cells expressing Gln1(R23E)-mCherry were spheroplasted and lysed in phosphate buffers of different pHs. Images were acquired immediately after lysis. The scale bar is 5 μm. Note that the lysis buffer did not contain a crowder. (B) Yeast cells expressing mCherry-tagged Gln1 were washed twice with water and resuspended in a glucose-containing buffer of the indicated pHs with or without the proton carrier 2,4-dinitrophenol (DNP). Images were taken 1 hr after addition of the buffer. The scale bar is 5 μm. (C) Yeast cells expressing Gln1-mCherry were washed twice with water and resuspended in phosphate buffers of different pHs to induce starvation. The buffers contained proton carriers and energy inhibitors for rapid equilibration of inside and outside pH. The scale bar is 5 μm. (D) Gln1-mCherry was purified from yeast and incubated in an acidic or basic buffer containing a crowding agent for 1 hr. Samples were analyzed by fluorescence microscopy. The scale bar is 3 μm.
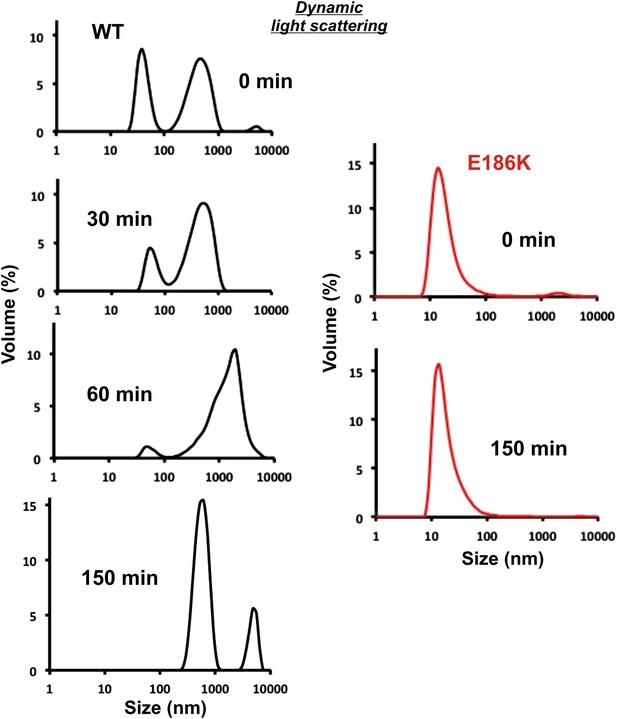
Equal amounts of 6xHis-tagged wild type and E186K Gln1 purified from bacteria were mixed with an acidic buffer and subjected to dynamic light scattering.
Shown is the volume distribution that was derived from the intensity distribution.
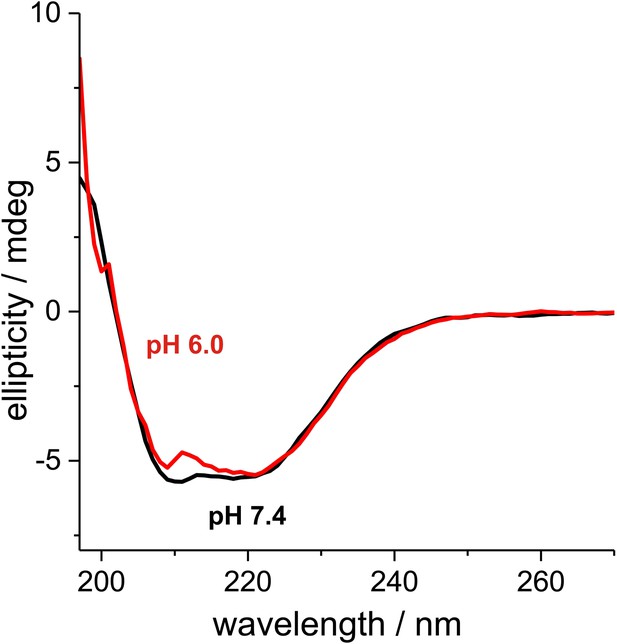
6xHis-tagged wild-type Gln1 purified from bacteria was subjected to Far-UV CD at pH 7.4 and 6.
The degree of helicity is essentially unchanged as evident from the almost unaffected 222 nm signal. This indicates that Gln1 retains a near-native structure in conditions that induce assembly into higher order structures.
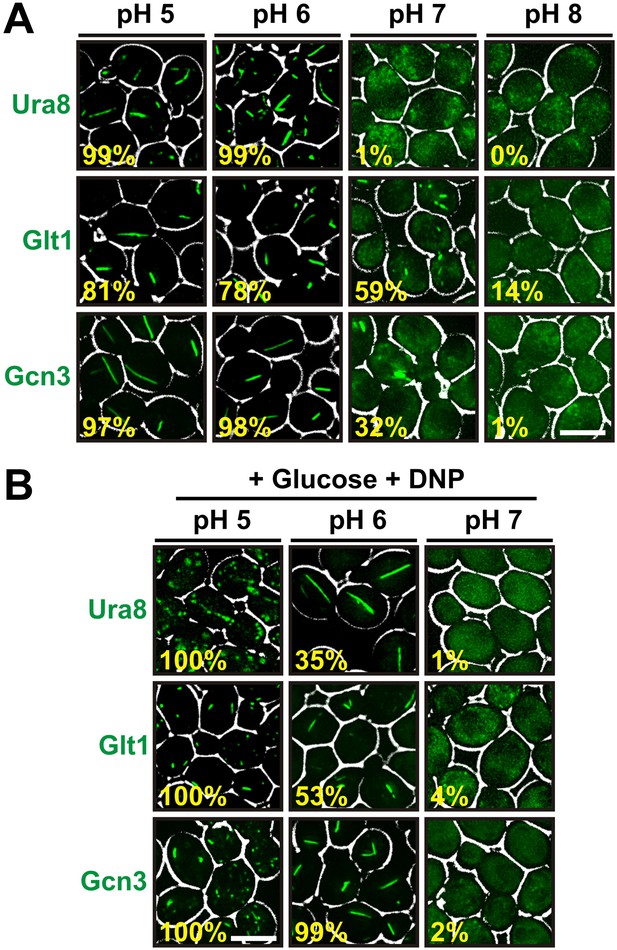
Other metabolic enzymes form filaments in a pH-dependent manner.
(A) Yeast cells expressing sfGFP(V206R)-tagged Ura8, Glt1, or Gcn3 were washed twice with water and resuspended in buffers of different pHs to induce starvation. Images were taken 2 hr after onset of starvation. The scale bar is 5 μm. (B) Yeast cells expressing sfGFP(V206R)-tagged Ura8, Glt1, or Gcn3 were washed twice with water and resuspended in a buffer containing 2% glucose and the proton carrier 2,4-dinitrophenol (DNP). Images were taken 1 hr after addition of the buffer. The numbers in yellow give the percentage of cells with fluorescent foci. At least 200 cells were counted. The scale bar is 5 μm.
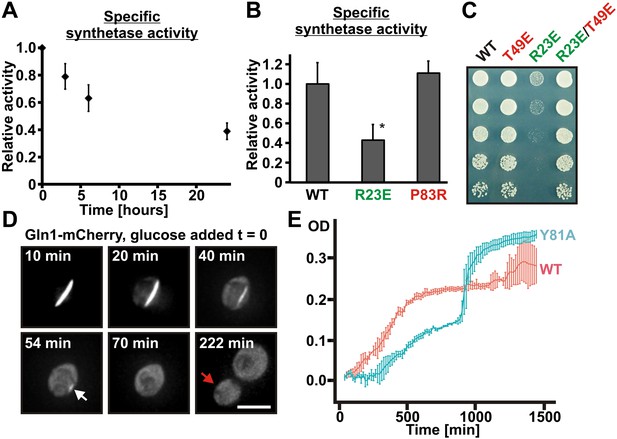
Assembled Gln1 is catalytically inactive but becomes active again after disassembly.
(A) Lysates were prepared form wild-type cells exposed to advanced starvation conditions for 3, 6, or 24 hr, and the glutamine synthetase activity was determined as described previously (Mitchell and Magasanik, 1984). The obtained values were normalized to the amount of Gln1 in the cell lysate based on immunoblotting experiments. The shown data is the mean of five biological replicates (±SEM). (B) Lysates were prepared from yeast cells expressing wiltype or variant Gln1-V5 and the glutamine synthetase activity was determined as in (A). The values were normalized to the amount of Gln1 contained in the lysate based on immunoblotting experiments (*p value <0.05). (C) Endogenous Gln1 was substituted with the indicated wild type or variant versions expressed from an ADH1 promoter-containing plasmid. Cells were grown over night and equal amounts of late log phase cells were spotted onto synthetic plates in serial 1:5 dilutions. (D) Cells expressing Gln1-mCherry from a GAL-inducible promoter were washed twice with water and starved in a phosphate–citrate buffer of pH 6 for 4 hr. Filament dissolution was followed after re-addition of glucose by time-lapse imaging. The red arrow denotes the newly formed bud. Note that filament dissolution and bud emergence take longer as the cells had to readjust to a new carbon source (glucose instead of galactose). The scale bar is 5 μm. Also see corresponding Video 5. (E) Wild-type and mutant (Y81A) yeast were exposed to advanced starvation conditions and regrowth was monitored after addition of nutrients (we used only 0.5% glucose in this assay as this led to slower regrowth and a more pronounced lag phase). Note that the Y81A cultures reached a slightly higher optical density, probably because of compensatory changes in the gene expression network. The values show the mean of four technical replicates. The data shown are representative of four independent experiments.
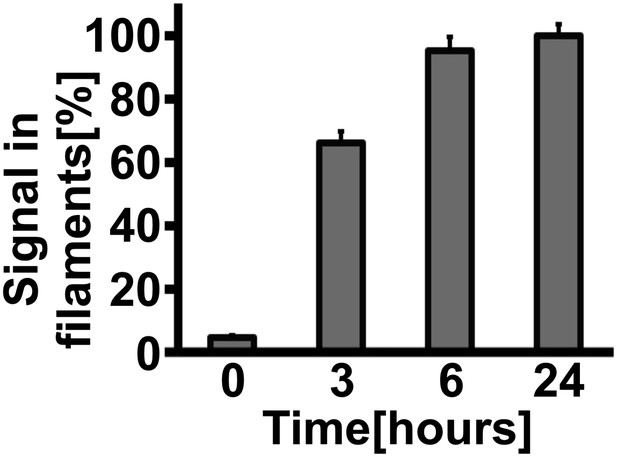
Gln1 filaments form progressively after exposure of yeast to advanced starvation conditions.
Cells expressing Gln1-mCherry were washed twice with water and resuspended in a phosphate–citrate buffer of pH 6 to induce starvation. Images were acquired 0, 3, 6, and 24 hr after onset of starvation. The total cellular mCherry signal was segmented into a diffuse and filament fraction using the Squassh segmentation tool from the Mosaic Suite of the Fiji image analysis software. The values shown are the mean (±SEM) of three independent experiments (p<0.05). The y-axis gives the fraction of the signal bound up in filaments. The values were normalized to the 24-hr time point as no further filament formation was observed beyond this time point.
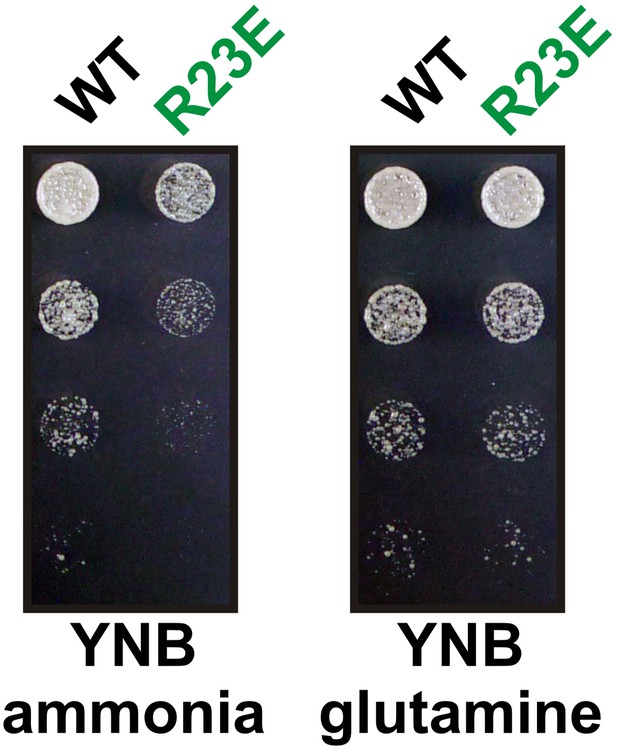
The R23E growth defect can be rescued by adding glutamine to the growth medium.
Endogenous Gln1 was deleted and substituted with wild-type or variant Gln1 expressed from a plasmid. Cells were grown over night and equal amounts of late log phase cells were spotted onto synthetic plates in serial 1:5 dilutions. The plate media lacked amino acids and contained ammonia or glutamine as the only nitrogen source. The strain background was a prototrophic variant of W303 (Klosinska et al., 2011).
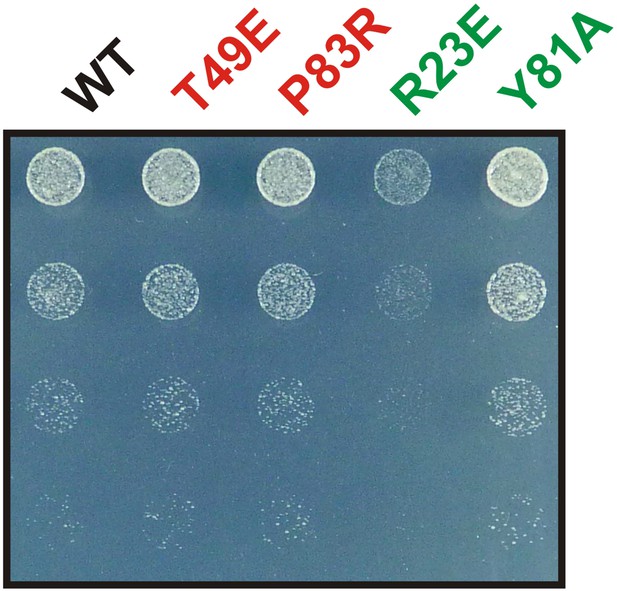
The Y81A mutation does not affect growth.
Endogenous Gln1 was substituted with the indicated wild-type or variant versions expressed from an ADH1 promoter-containing plasmid. Cells were grown over night and equal amounts of late log phase cells were spotted onto synthetic plates in serial 1:5 dilutions.
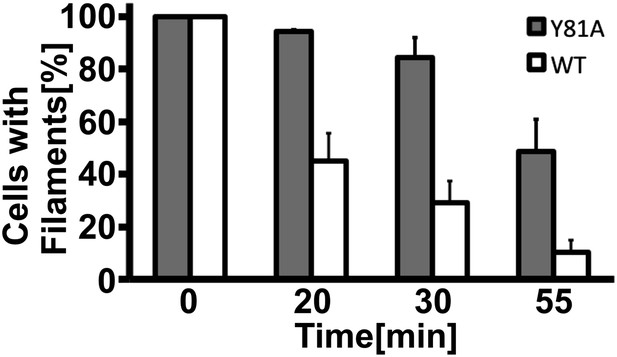
Filament dissolution is impaired in Y81A mutants.
Cells expressing WT or Y81A Gln1-mCherry were washed twice with water and resuspended in a phosphate-citrate buffer of pH 6 to induce starvation. After 4 hr of starvation, nutrients were resupplied and images were acquired 0, 20, 30, and 55 min. The y-axis gives the fraction of cells containing filaments. Mean values are shown (±SEM).
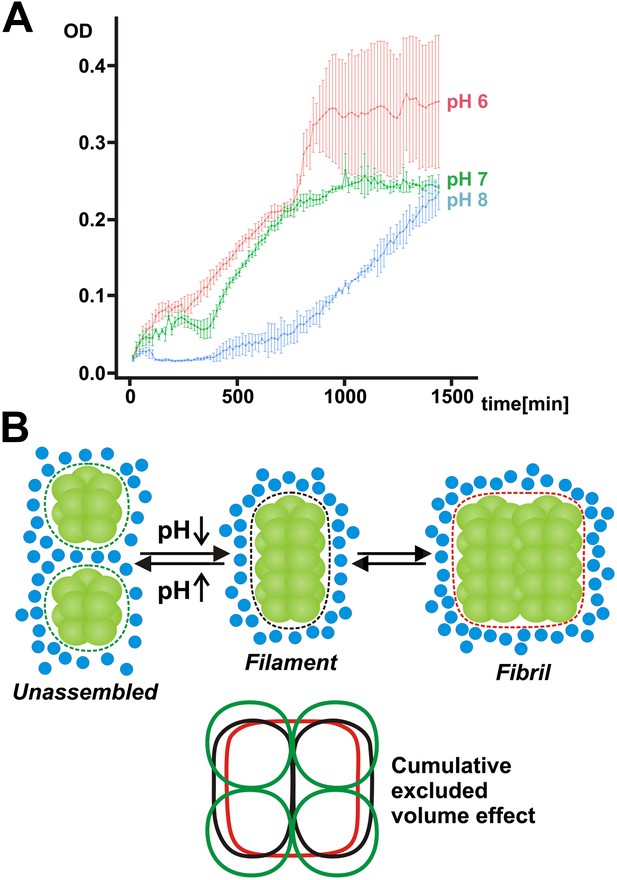
Mechanism of filament formation by Gln1 and its potential role in starvation survival.
(A) Acidification of the yeast cytosol promotes survival and recovery from starvation. Yeast cells were exposed to advanced starvation conditions using buffers with a pH of 6, 7 or 8 for 3 days. Upon re-addition of SD medium containing a limited amount of glucose (0.5%), regrowth was monitored in a plate reader. The values show the mean of four technical replicates (±SEM). The data shown are representative of four independent experiments. (B) Mechanism of pH-induced filament formation by metabolic enzymes. Gln1 assembles into filaments by a back-to-back enzyme stacking mechanism. Transitions between unassembled Gln1 decamers (left), filaments (middle), and fibrils (right) are driven by pH changes and excluded volume effects. Assembly requires an acidic pH, whereas disassembly requires a basic pH. Gln1 enzyme complexes are shown in green. Blue spheres denote inert macromolecules that are excluded from the space that is occupied by Gln1. This inaccessible space is indicated by the dotted lines. The bottom diagram illustrates the cumulative excluded volume effect that entropically drives filament assembly and fibril formation. The colors of the bottom diagram correspond to the colors of the excluded volume areas above.
Videos
Gln1 forms filaments in starved yeast. Cells expressing Gln1-mCherry were washed twice with water and resuspended in a phosphate-citrate buffer of pH 6 to induce starvation (time point 0). Filament formation was followed by time-lapse microscopy. Time points are indicated in minutes.
Gln1 forms filaments in starved yeast. Cells expressing Gln1-mCherry were washed twice with water and resuspended in a phosphate-citrate buffer of pH 6 to induce starvation (time point 0). Filament formation was followed by time-lapse microscopy. Time points are indicated in minutes.
Gln1 filaments dissolve upon glucose addition to starved cells. Cells expressing Gln1-mCherry were washed twice with water and resuspended in a phosphate-citrate buffer of pH 6 to induce starvation (time point 0). The cells were incubated for 4 hr to induce the formation of filaments. At time point 0, glucose (2%) was added and filament dissolution was followed by time-lapse microscopy.
Gln1 filaments dissolve upon glucose addition to starved cells. Cells expressing Gln1-mCherry were washed twice with water and resuspended in a phosphate-citrate buffer of pH 6 to induce starvation (time point 0). The cells were incubated for 4 hr to induce the formation of filaments. At time point 0, glucose (2%) was added and filament dissolution was followed by time-lapse microscopy.
Filamentous Gln1 can be reactivated upon entry into the cell cycle. Cells expressing Gln1-mCherry from a GAL-inducible promoter were washed twice with water and starved in a phosphate-citrate buffer of pH 6 for 4 hr. At time point 0, glucose (2%) was added and filament dissolution was followed by time-lapse microscopy. Note that filament disassembly and bud emergence take longer, because the cells had to readjust to a new carbon source (glucose instead of galactose).
Additional files
-
Supplementary file 1
(A) Plasmids used in this study. (B) Antibodies used in this study. (C) Yeast strains used in this study.
- https://doi.org/10.7554/eLife.02409.034





