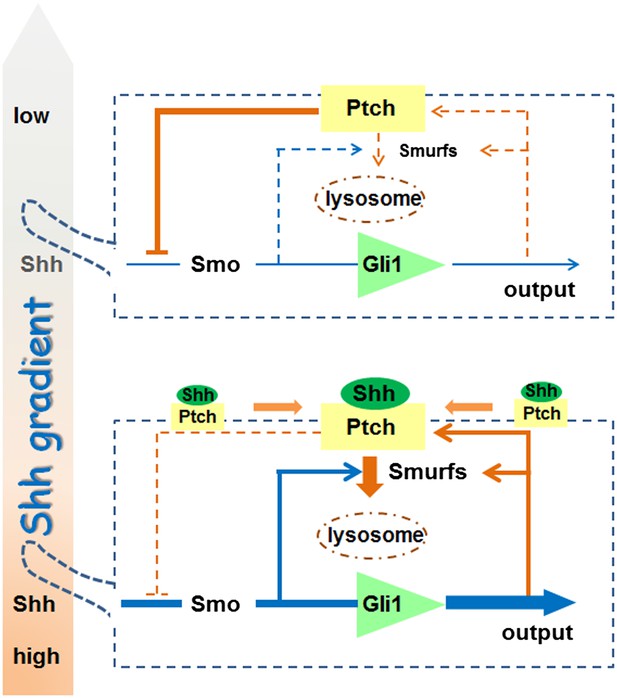
Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception
Figures
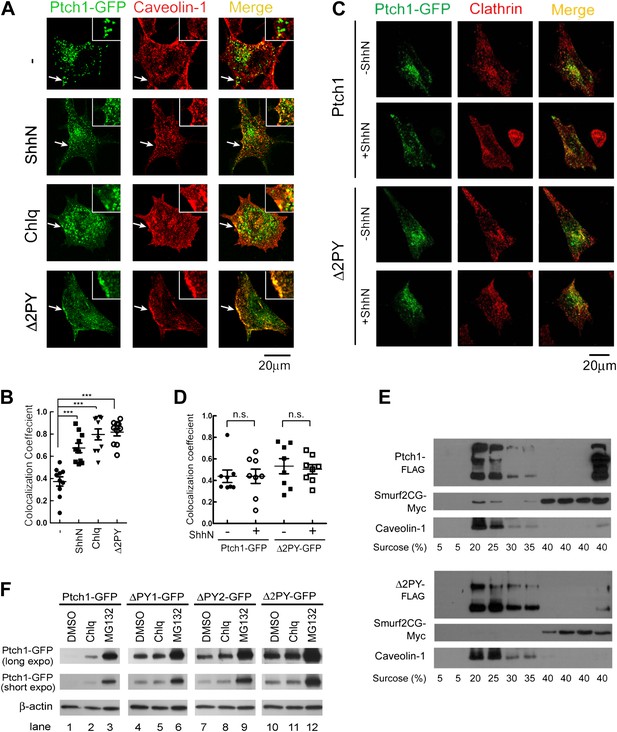
The PPXY motifs define sorting signals from lipid rafts to late endosomes.
(A) Confocal images showing colocalization of exogenously expressed Ptch1-GFP or Δ2PY (green) with native Cav-1 (red) at the rim of the plasma membrane, and (B) calculation of the colocalization coefficients in (A) in transfected MEFs. ShhN-CM and Chlq were added 1 hr prior to fixation. The chamber slides were chilled at 4°C for 20 min and then shifted to 37°C for another 20 min before fixation with 4% paraformaldehyde in PBS. The cells were then permeabilized with 0.5% Triton X-100 and stained with an antibody for Cav-1. (C) Representative images and (D) quantification of colocalization between Ptch1-GFP and clathrin heavy chain. MEFs were transfected with Ptch1-GFP or PtchΔ2PY-GFP, then treated with ShhN or Ctrl medium for 1 hr before fixation. (E) Western analyses of sucrose gradient fractions showing Ptch1-FLAG co-sedimented with Smurf2CG-Myc and Cav-1. Δ2PY was inefficient in bringing Smurf2CG-Myc into Cav-1 positive sedimentation fractions. (F) Western blot analyses of stabilities of Ptch1 and the ‘PPXY’ motif mutants in MEFs. Chlq or MG132 treatment was carried out for 4 hr. The confocal images were taken with a 63x objective, and the insets in 1A were digitally magnified. Bars represent mean ± standard deviation (SD). Statistical analyses were performed by two-tail Student's t test. ***p<0.001, and n.s., not statistically significant (p>0.05).
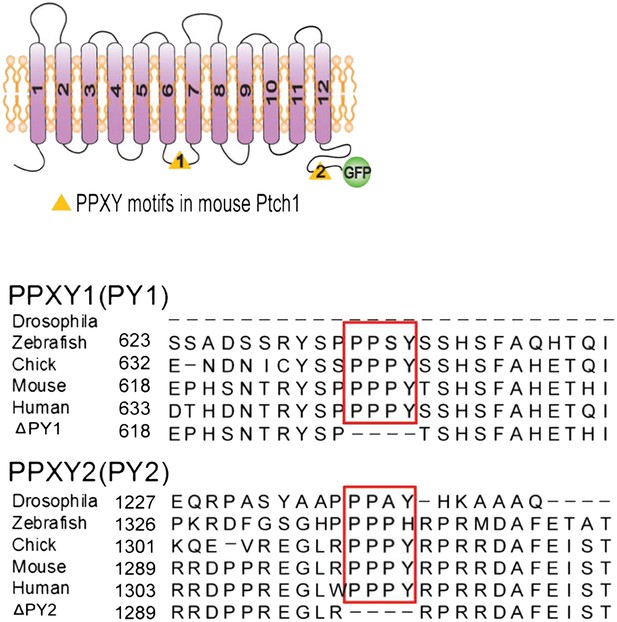
Position and sequence alignment of ‘PPXY’ motifs.
(A) Schematic representation of Ptch1 constructs (left) and sequence alignments (right) of Drosophila and vertebrate Ptch1 surrounding the two evolutionarily conserved PPXY motifs.
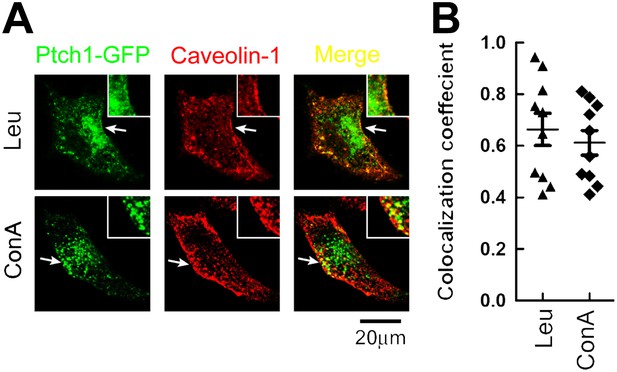
Lysosomal inhibitors cause Ptch1-GFP to accumulate in lipid rafts.
(A) Representative confocal images showing accumulation of Ptch1-GFP in Cav-1 positive lipid rafts after blocking endocytosis with lysosomal inhibitors Leu and ConA. (B) Calculation of colocalization coefficients in (A). The confocal images were taken with a 63x oil lens, and the insets were digitally magnified. Bars represent mean ± standard deviation (SD).
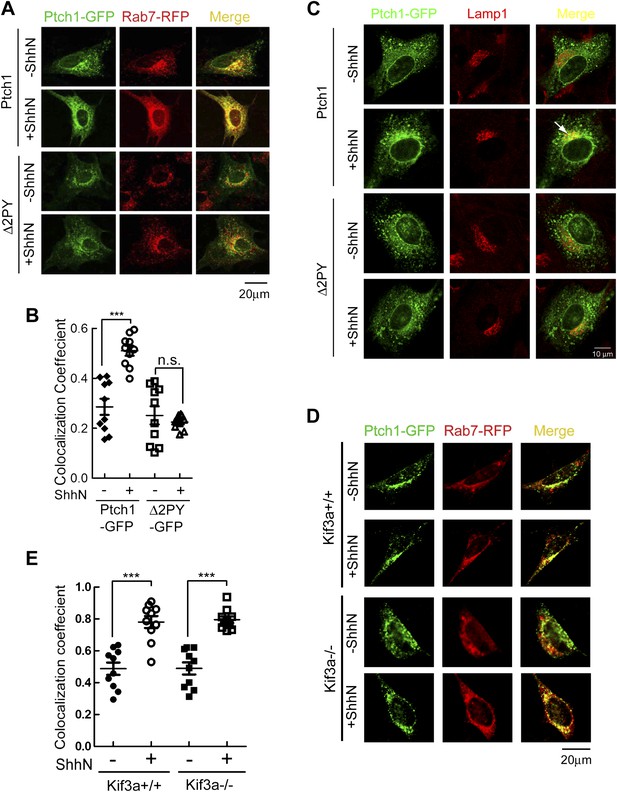
PPXY motifs are required for Shh-induced endocytosis of Ptch1.
(A) Confocal images showing colocalization of Ptch1-GFP or Δ2PY (green) with Rab7-RFP (red), and (B) calculation of the colocalization coefficients in (A) in transfected MEFs. (C) Confocal images showing localization of Ptch1-GFP or Δ2PY (green) in vesicles marked anti-Lamp1 (red) in the presence of 1 mg/ml leupeptin. (D) Confocal imaging and (E) calculation of colocalization coefficient of Ptch1-GFP and Rab7-RFP in Kif3a−/− and control MEFs. ShhN treatment was for 1 hr and the cells were processed as in Figure 1A. Statistical analyses were performed by two-tail Student's t test. ***p<0.001, and n.s., not statistically significant (p>0.05).
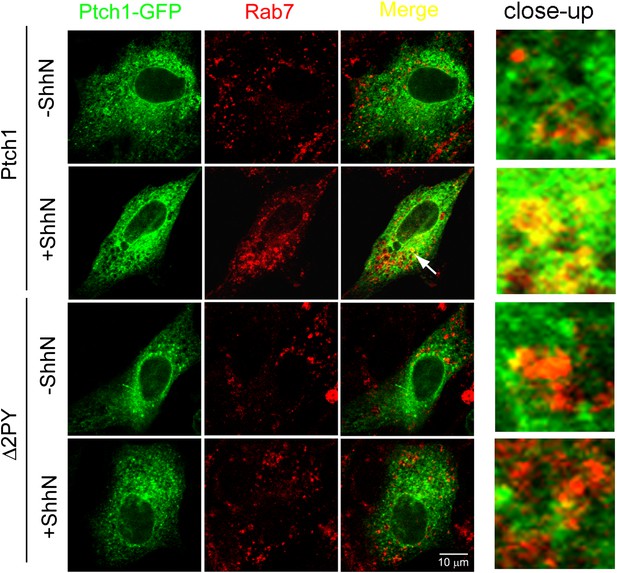
Shh promotes colocalizaiton of Ptch1-GFP with endogenous Rab7 in late endosomes.
Representative confocal images showing ShhN treatment promotes colocalization of Ptch1-GFP in late endosomes visualized by anti-Rab7. Transfected MEFs were treated with ShhN-CM or control conditioned medium for 1 hr, followed by incubations at 4°C for 20 min and 37°C for 20 min. The close-up images were digitally amplified.
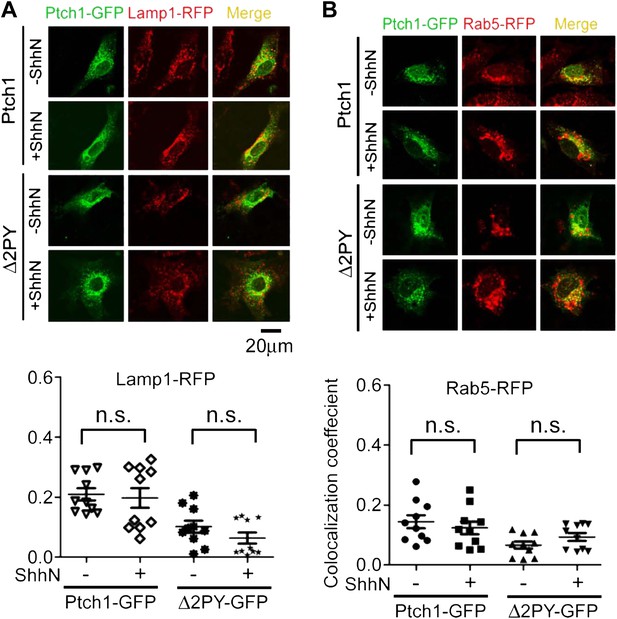
Lack of colocalization of Ptch1-GFP or Δ2PY-GFP with exogenous Rab5-RFP and Lamp1-RFP without blocking lysosomal turnover.
Representative Confocal images and quantification of colocalization coefficients showing that Ptch1-GFP or Δ2PY-GFP does not colocalize with Lamp1-RFP (red) (A) or Rab5-RFP (B). Transfected MEFs were treated ShhN or control conditioned medium without Leupeptin for 2 hr, and then the cells were processed as in Figure 2A.
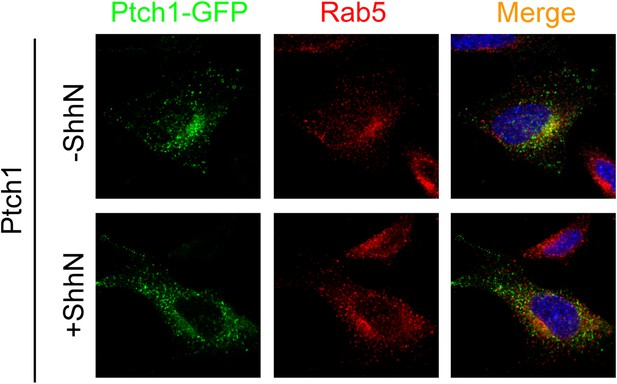
Ptch1-GFP or Δ2PY-GFP was not found in early endosomes marked by anti-Rab5 immunofluorescence staining.
Representative Confocal images showing Ptch1-GFP or Δ2PY-GFP and endogenous Rab5 in non-overlapping green or red channel, respectively.
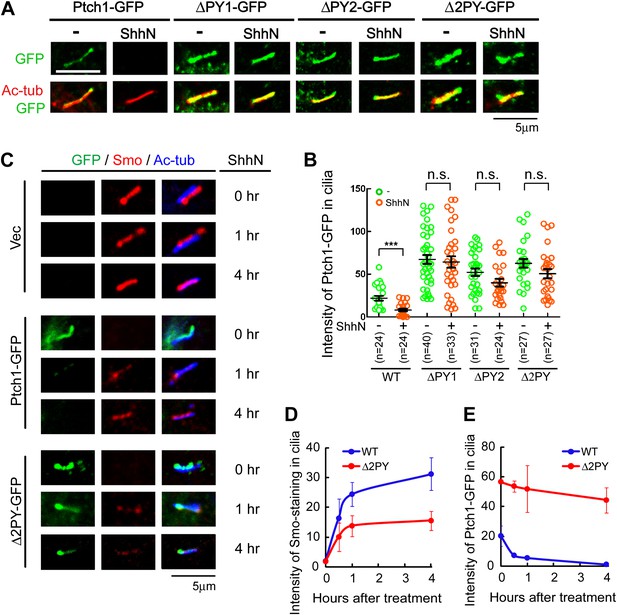
The ‘PPXY’ motifs regulate the opposing movements of Ptch1 out of and Smo into the primary cilium.
(A) Representative confocal images and (B) distribution of GFP fluorescence showing accumulation of the ‘PPXY’ motif mutants of Ptch1 in primary cilia in the absence or presence of ShhN. Two-tail Student's t test was used for statistical analysis. ***p<0.001, n.s., not significant (p>0.05). (C) Immunofluorescence of GFP as well as endogenous Smo (red) and acetylated tubulin (blue) staining in Ptch1−/− MEFs transfected with Ptch1-GFP or Δ2PY. (D) Quantification of anti-Smo staining and (E) GFP fluorescence as in (C). Only transfected GFP positive cells were counted for the ciliary localization of endogenous Smo. In all of the above experiments, transfected cells were grown to confluence and then serum-starved for 24 hr to allow for ciliogenesis. ShhN-CM treatment was for 24 hr, or as indicated.
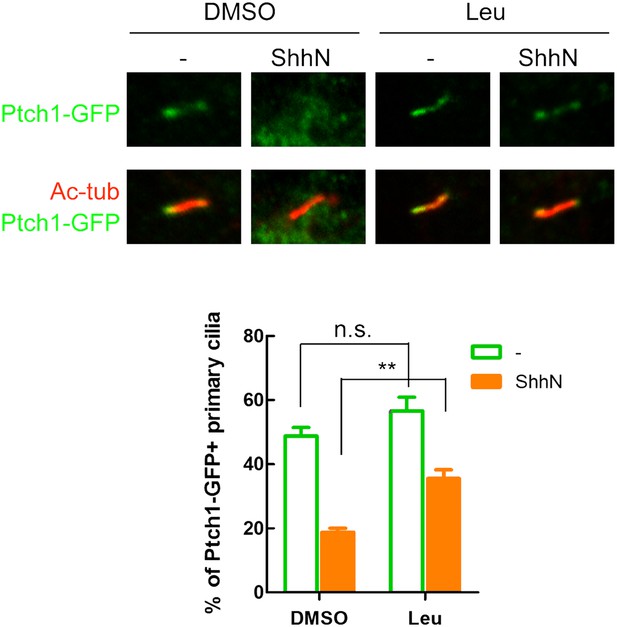
Inhibition of Lysosomal turnover dampens Shh-induced ciliary exit of Ptch1-GFP.
Representative confocal images and calculations thereof showing Ptch1-GFP fluorescence accumulated in primary cilia. ShhN and leupeptin (1 mg/ml) were added to the WT MEFs for 2 hr. Two-tail Student's t test was used for statistical analysis. **p<0.01, n.s., not significant (p>0.05).
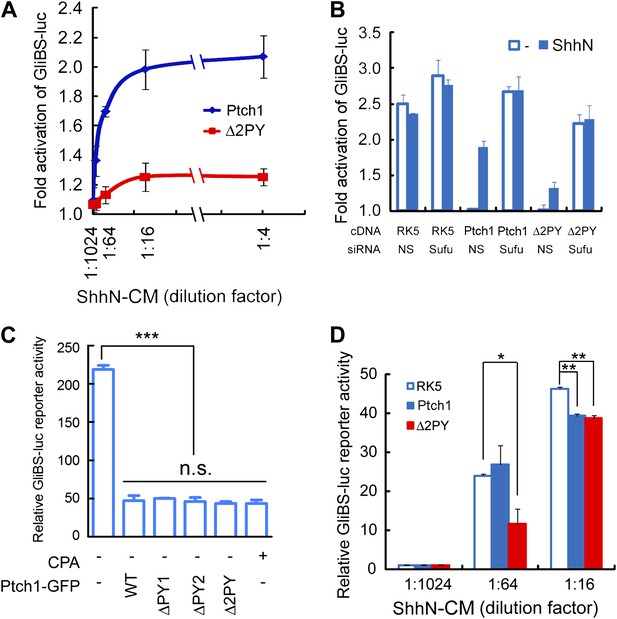
The ‘PPXY’ motifs are required for eliciting both cell and non-cell autonomous transcriptional responses to Shh.
(A) Luciferase assays for Ptch1 and the Δ2PY mutant in Ptch1−/− MEFs that were transfected together with the 8xGliBS-luc reporter construct. Each data point was obtained in triplicate and the error bars denote the standard error. (B) Rescuing Shh induction blockade imposed by Δ2PY using siSufu in Ptch1−/− MEFs. The experiment was set up as in (A) except that Sufu was knocked down by siRNA at the same time as cDNA transfection and 1:16 dilution of ShhN-CM was used. (C) Relative activities of the GliBS-luc reporter that was co-expressed with Ptch1 or the ΔPY mutants in Ptch1−/− MEFs without ShhN-CM treatment. The ΔPY mutants displayed same inhibitory effect as WT Ptch1. (D) Non-cell autonomous inhibition of GliBS-luc reporter in neighboring cells. Ptch1−/− MEFs transfected with Ptch1-FLAG, Δ2PY, or the vector control were mixed at 5:1 ratio with NIH3T3:GliBS-luc reporter cells. The cells were given ShhN-CM for 24 hr, and two-tail Student's t test was used for statistical analyses. *p<0.05, **p<0.01, ***p<0.001, and n.s., not significant (p>0.05).
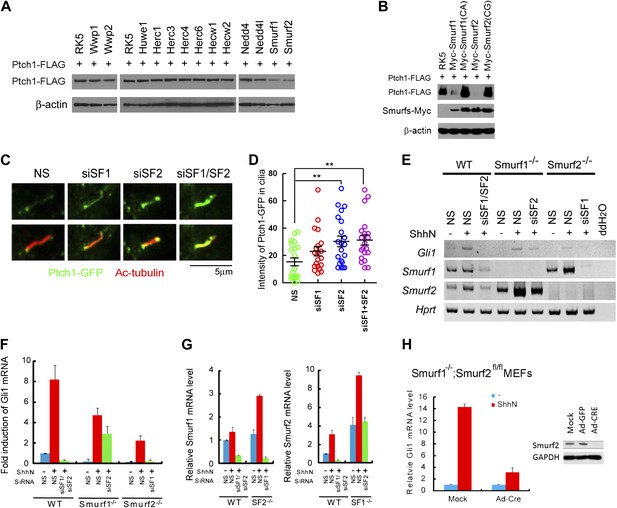
Smurf1 and Smurf2 are E3 ligases required for Shh signaling.
(A) Western analyses of Ptch1-FLAG in HEK293 cells that were co-transfected with cDNAs encoding a battery of HECT-domain E3 ligases as indicated, and (B) ligase deficient Smurf mutants. β-actin was used as a loading control. (C) Representative confocal images and (D) calculations of Ptch1-GFP fluorescence accumulated in primary cilia as the result of siRNA knockdown of Smurf1, Smurf2, or both in NIH3T3 cells. Primary cilia were marked by acetylated tubulin (red). (E) RT-PCR detection of Gli1, Smurf1, and Smurf2 mRNAs in wildtype (WT), Smurf1−/− , and Smurf2−/− MEFs transfected with non-silencing (NS) or Smurf-specific siRNAs as indicated. HPRT mRNA was used as an internal control. A representative gel image is shown here. (F) RT-qPCR quantification of fold induction of Gli1 mRNA from an experiment as in (E). Fold induction was calculated using Gli1 mRNA level normalized against that of Hprt for even loading and then against the normalized Gli1 mRNA level from cells transfected with NS siRNA and without ShhN treatment. (G) RT-qPCR analysis of relative levels of Smurf1 and Smurf2 mRNAs from the experiment in (F). (H) RT-qPCR detection of endogenous Gli1 mRNAs in Smurf1−/−;Smurf2fl/fl MEFs that were infected with either Ad-GFP (mock) or Ad-Cre for 12 hr, and then treated with either control or ShhN conditional medium for 72 hr. (I) Western analyses of endogenous Smurf2 in Smurf1−/−;Smurf2fl/fl MEFs from the experiment in (H).
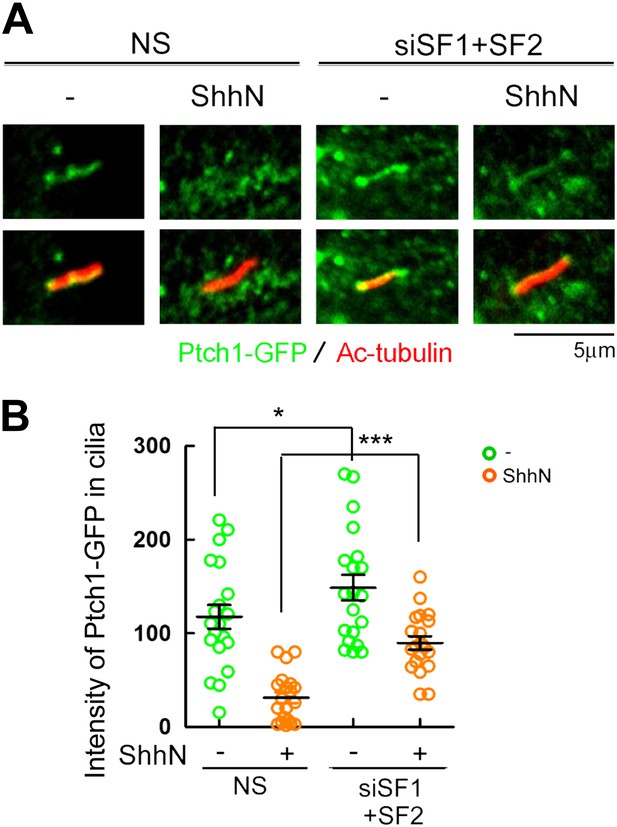
Knockdown of Smurf1 and Smurf2 simultaneously dampens Shh-induced ciliary exit of Ptch1-GFP.
Representative confocal images and calculations thereof showing Ptch1-GFP fluorescence accumulated in primary cilia. NIH3T3 cells were transfected with siRNAs specific for Smurf1 and Smurf2, and then the cells were treated with control or ShhN conditioned medium before Ptch1-GFP was visualized in cilia and quantified. Two-tail Student's t test was used for statistical analysis. *p<0.05, ***p<0.001.
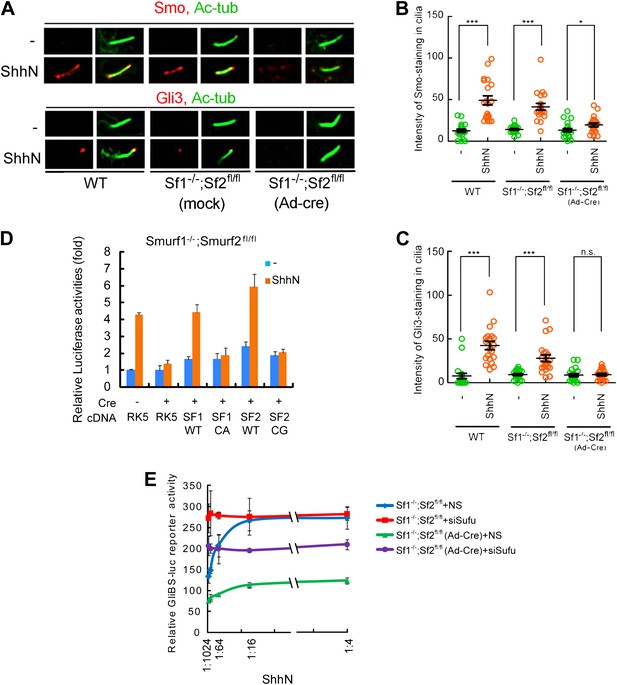
Smurf1 and Smurf2 are required For Shh signaling.
(A) Representative confocal images of Smo and Gli3 immunofluorescence staining in cilia of wildtype (WT), Smurf1−/−;Smurf2fl/fl, or Smurf1−/−;Smurf2fl/fl MEFs infected with Ad-Cre viruses. (B) Quantification of Smo and (C) Gli3 immunofluorescence staining in cilia of (A). In the above experiments, ShhN treatment was carried out for 24 hr, and the means and standard deviation were calculated from two independent experiments (n = 20). (D) GliBS-luc assays in Smurf1−/−;Smurf2fl/fl MEFs showing the deficiency of Shh induction associated with genomic ablation of both Smurfs can be rescued by re-introducing either wildtype Smurf1 or Smurf2 but not their corresponding mutants. (E) GliBS-luc reporter assays for the ability of siSufu to by-pass the requirement of Smurfs in Shh signaling. Smurf1−/−;Smurf2fl/fl MEFs were infected with Ad-cre and then transfected with siSufu or ns control. The cells were then treated with a series of dilutions of ShhN-CM before luciferase activities were assayed. Error bars denote standard deviations. Statistical analyses were performed by two-tail Student's t test. *p<0.05, **p<0.01, ***p<0.001, and n.s., not significant (p>0.05).
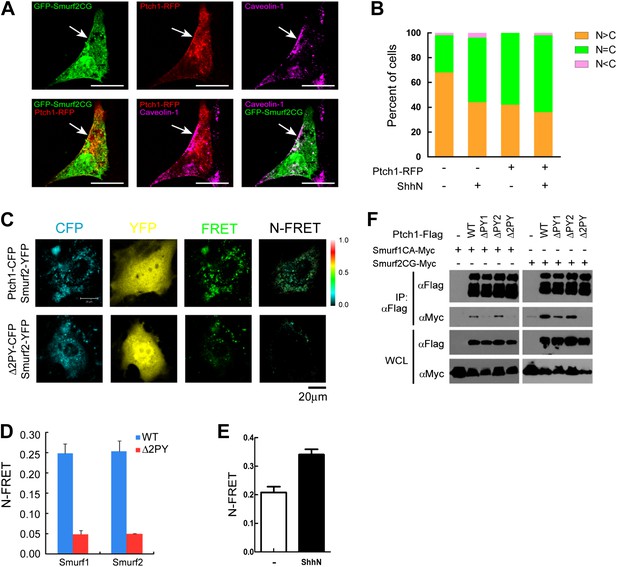
Colocalization and interaction between Ptch1 and Smurfs in Cav-1 positive lipid rafts.
(A) Confocal images showing colocalization of GFP-Smurf2CG and Ptch1-RFP in Cav-1 positive lipid rafts. The cells were not permeabilized before they were stained with anti-Cav-1, and the images were taken with a 63x oil lens. (B) Quantification of nuclear and cytoplasmic distribution of Smurf2GFP as in Figure 7—figure supplement 1. The percentage of mostly nuclear (N > C), even distribution (N = C), or mostly cytoplasmic (N < C) of Smurf2GFP pattern cells was calculated based images of 40 cells at each data point. (C) FRET analysis of Ptch1-CFP or Δ2PY-CFP interaction with Smurf1-YFP or Smurf2-YFP in transfected MEFs. Representative images of CFP, YFP, FRET fluorescence, and N-FRET are shown. (D) Quantification of N-FRET values using the sensitized emission method, which is expressed as means plus SD in the bar graph. (E) FRET analysis of Ptch1-CFP interaction with Smurf2-YFP in transfected MEFs that were treated with ShhN or control conditioned medium for 2 hr. Quantification of N-FRET values described in (D). (F) Co-immunoprecipitation analyses of FLAG-Ptch1 and the ‘PPXY’ motif mutants with Myc-tagged Smurf1CA or Smurf2CG ligase-deficient mutants. The immunocomplexes were precipitated using anti-FLAG, and blotted with anti-Myc.
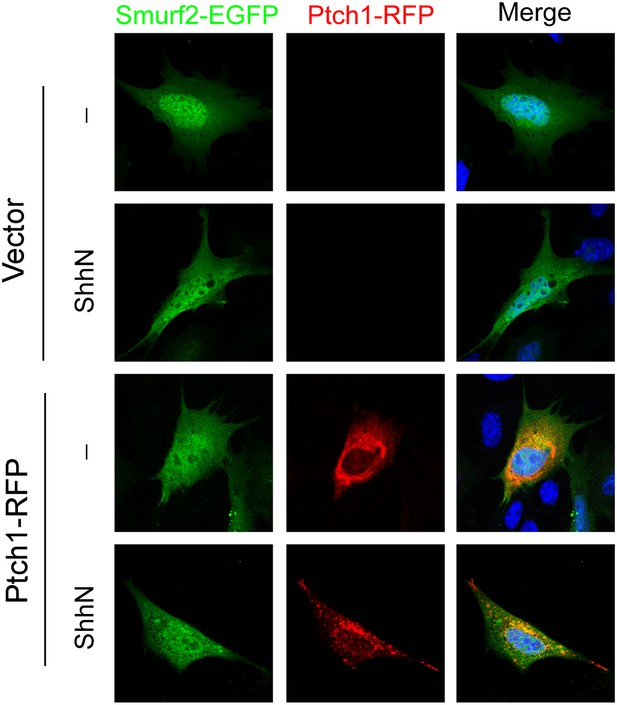
ShhN treatment and co-expression with Ptch1 caused Smurf2 to redistribute from the nucleus to the cytoplasm.
Representative fluorescent images showing subcellular localization of Smurf2GFP in MEFs co-transfected with empty vector or Ptch1-RFP and treated without or with ShhN conditioned medium.
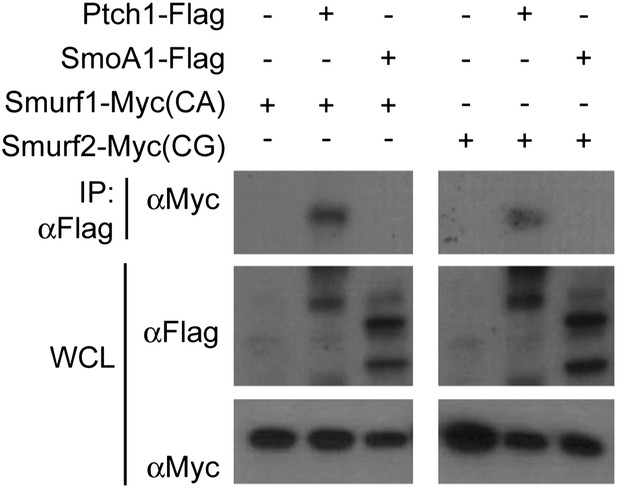
Neither Smurf1 nor Smurf2 interact with Smo.
Western analyses of Ptch1-FLAG or SmoA1-FLAG immunoprecipitated with anti-FLAG in HEK293 cells that were also co-transfected with Myc-tagged Smurf1CA or Smurf2CG ligase-deficient mutants. Although Smurf1CA or Smurf2CG was readily detected in the anti-Ptch1-FLAG precipitates, they did not co-precipitate with Smo-FLAG.
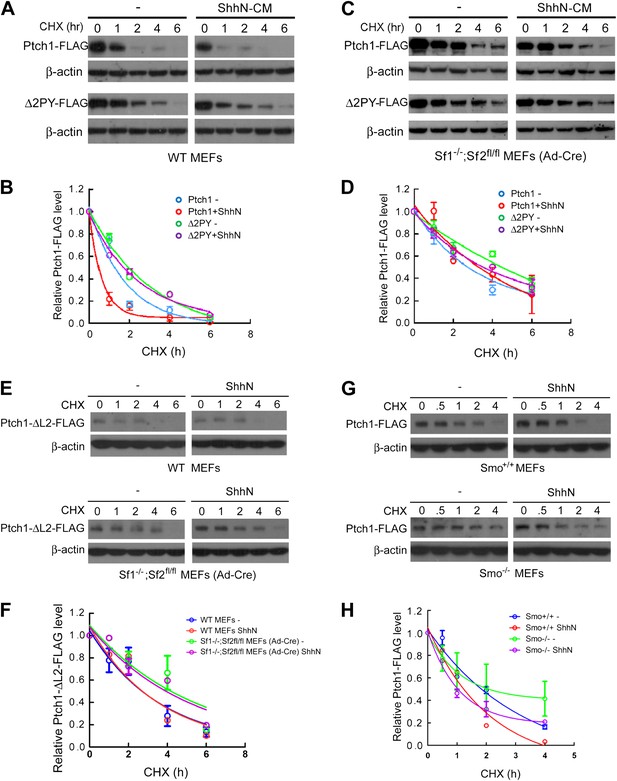
Smurfs are required for the Shh-induced endocytic turnover of Ptch1.
Western analysis of Ptch1-FLAG and Δ2PY-FLAG turnover rates (A) and quantification thereof (B) in WT MEFs. ShhN and CHX were added for duration as indicated. (C) Western analysis of Ptch1-FLAG and Δ2PY-FLAG turnover rates (C) and quantification thereof (D) in Smurf1−/−;Smurf2fl/fl MEFs infected with Ad-cre. (E) Western analysis of Ptch1-Δloop2-FLAG turnover rate and quantification thereof (F) in WT (upper) and Smurf1−/−;Smurf2fl/fl MEFs infected with Ad-cre (lower). (G) Western analysis of Ptch1-FLAG turnover rate and quantification thereof (H) in WT (upper) and Smo−/− MEFs (lower). Each data point denotes mean ± standard deviation from two independent experiments.
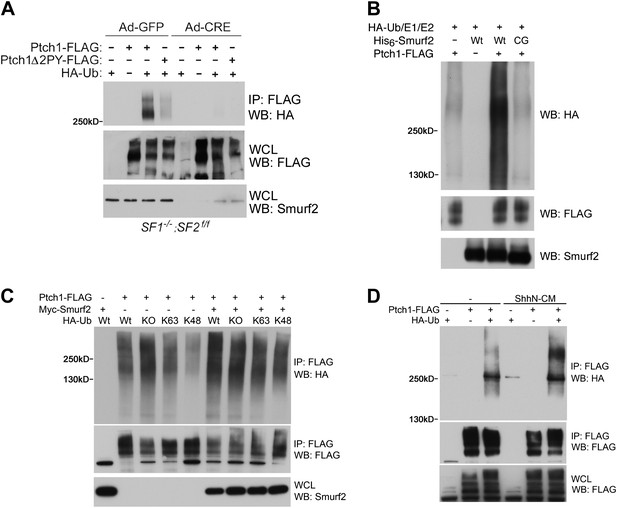
Smurfs are required for ubiquitin modification of Ptch1.
(A) Western analysis of ubiquitinated Ptch1-FLAG and Ptch1Δ2PY-FLAG in Smurf1−/−;Smurf2fl/fl MEFs infected with Ad-GFP or Ad-Cre. These MEFs were first infected with adenoviruses and then transfected with HA-Ub and the Ptch1 plasmids as marked. The exogenously expressed Ptch1 proteins were immunoisolated using anti-FLAG beads prior to analysis. (B) Western analysis of Ptch1-FLAG ubiquitination in vitro in a reconstituted system comprising purified recombinant His6-Smurf2 or the ligase-deficient His6-Smurf2CG from the baculovirus, HA-Ub, and an ATP regeneration system. Ptch1-FLAG was immunoisolated from HEK293 cells and the ubiquitination reaction was carried out on beads. The proteins were eluted prior to Western blot analysis. (C) Western analysis of ubiquitinated Ptch1-FLAG in Smurf2−/− MEFs that were also transfected with Wt, KO, K48, or K63 ubiquitin in the absence or presence of Myc-Smurf2. (D) Western analysis of ubiquitinated Ptch1-FLAG in WT MEFs treated with ShhN or control conditioned medium. Ptch1-FLAG in A-C was resolved by 6% SDS-PAGE, but a 4–12% gradient gel was used in D.
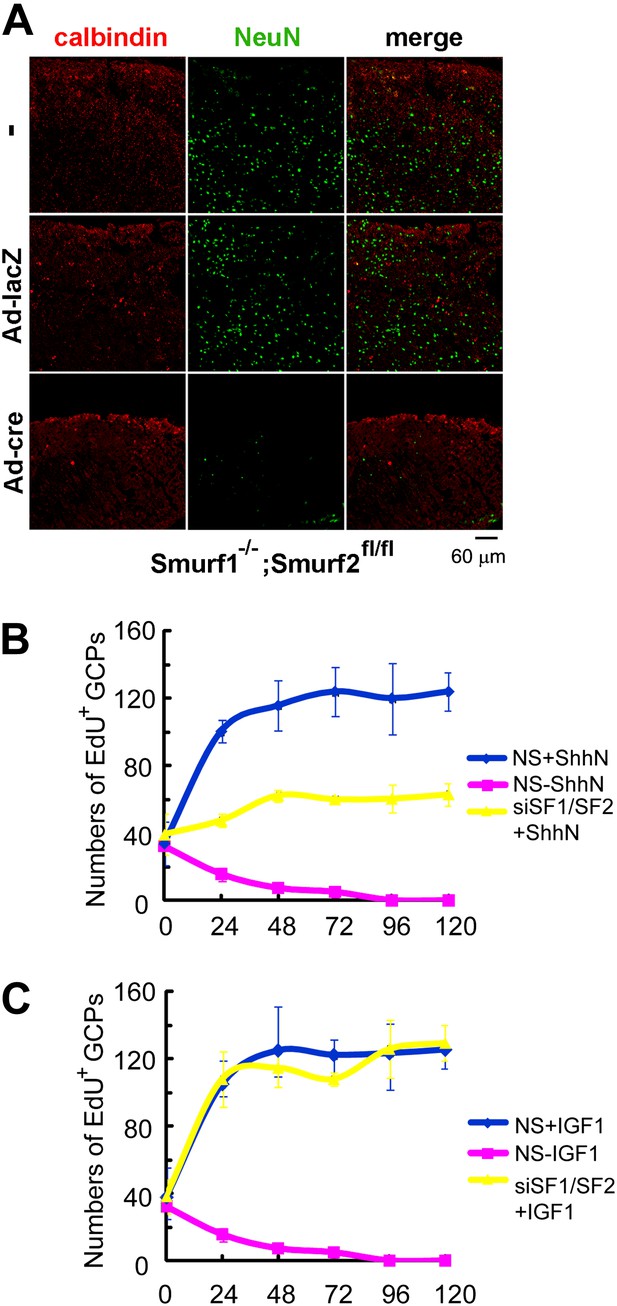
Requirements of Smurfs for Shh-induced organogenesis.
(A) Immunostaining of P7 cerebellar slices cultured in vitro with anti-calbindin (red) and anti-NeuN (green). The slices were first infected with control or cre-expressing adenoviruses for 24 hr and then continuously cultured for 12 days. Quantification of EdU incorporated GCPs in cerebellar slices cultured in the presence of ShhN from Figure 10—figure supplement 1A (B) or IGF-1 from Figure 10—figure supplement 1B (C), respectively. The data at each time point were derived from four separate fields, and the bars denote standard deviation.
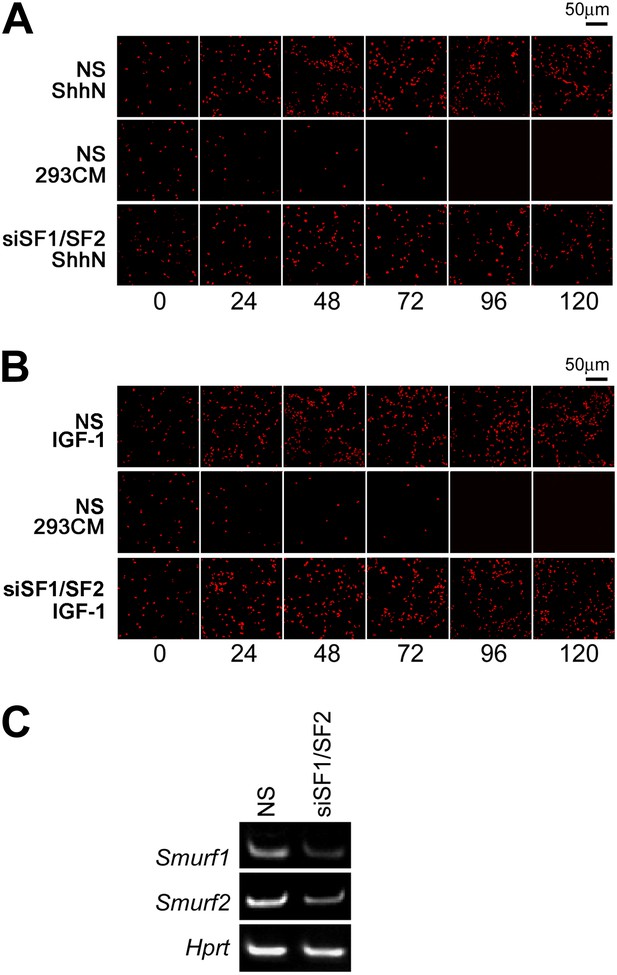
Smurfs are required for ShhN but not IGF-1 induced GCP proliferation.
EdU incorporation by GCPs growing in medium containing (A) ShhN or (B) IGF-1. Freshly isolated GCPs from normal C57/B6 mice were seeded in chamber slides that were coated with poly-D-lysine and Matrigel. The cells were then transfected with non-silencing (NS) control or Smurf1- and Smurf2-specific siRNAs. 12 hr later, Shh-N or IGF-1 conditioned medium was added to the culture and began the time point 0 hr. EdU was given to cells for 12 hr. (C) RT-PCR detection of Smurf1 and Smurf2 mRNAs for monitoring the siRNA knockdown efficiency.