Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2
Figures
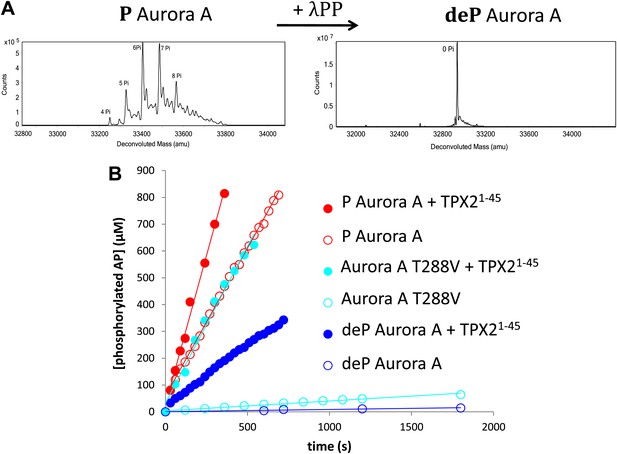
TPX21−45 drastically accelerates the kinetics of the dephosphorylated form of Aurora A kinase.
(A) Mass spectrometry data of heavily phosphorylated (P) and dephosphorylated (deP) Aurora A. The dephosphorylated protein was obtained after treatment of heavily phosphorylated, Escherichia coli-produced Aurora A with λ-protein phosphatase (λPP). (B) AP phosphorylation by dephosphorylated ( , 0.01 ± 0.005 s−1) or T288V mutant Aurora A (
, 0.01 ± 0.005 s−1) or T288V mutant Aurora A ( , 0.05 ± 0.002 s−1) is increased by up to 50-fold (
, 0.05 ± 0.002 s−1) is increased by up to 50-fold ( , 0.5 ± 0.1 s−1) and 25-fold (
, 0.5 ± 0.1 s−1) and 25-fold ( , 1.2 ± 0.1 s−1), respectively, in the presence of TPX21−45. This rate is comparable to the kinetics of phosphorylated Aurora A in the absence of TPX21−45 (
, 1.2 ± 0.1 s−1), respectively, in the presence of TPX21−45. This rate is comparable to the kinetics of phosphorylated Aurora A in the absence of TPX21−45 ( , 1.0 ± 0.2 s−1). Phosphorylated Aurora A shows up to a twofold increase in AP kinetics in the presence of TPX21−45 (
, 1.0 ± 0.2 s−1). Phosphorylated Aurora A shows up to a twofold increase in AP kinetics in the presence of TPX21−45 ( , 2.3 ± 0.2 s−1). Reactions are carried out in the presence of 1 μM protein, 50 μM TPX21−45, 5 mM ATP, and 1 mM AP in assay buffer (20 mM TrisHCl, 200 mM NaCl, 20 mM MgCl2, 3% (vol/vol) glycerol, 1 mM TCEP, pH 7.50) at 25°C. Phosphorylated peptide production was monitored by reverse phase-high performance liquid chromatography (RP-HPLC).
, 2.3 ± 0.2 s−1). Reactions are carried out in the presence of 1 μM protein, 50 μM TPX21−45, 5 mM ATP, and 1 mM AP in assay buffer (20 mM TrisHCl, 200 mM NaCl, 20 mM MgCl2, 3% (vol/vol) glycerol, 1 mM TCEP, pH 7.50) at 25°C. Phosphorylated peptide production was monitored by reverse phase-high performance liquid chromatography (RP-HPLC).
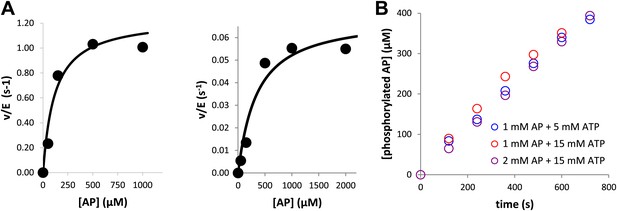
Kinase assays were conducted under saturating conditions of peptide and ATP.
To ensure peptide and nucleotide would not be rate-limiting, (A) the KM for AP was determined to be 116 ± 70 μM for phosphorylated Aurora A (left) and 320 ± 150 μM for dephosphorylated-mimic, T288V mutant Aurora A (right) and (B) the kinetics of 10 μM Aurora A T288V were monitored under 1 mM AP at 5 mM ATP ( , 0.0054 s−1) and 15 mM ATP (
, 0.0054 s−1) and 15 mM ATP ( , 0.0055 s−1) as well as 2 mM AP and 15 mM ATP (
, 0.0055 s−1) as well as 2 mM AP and 15 mM ATP ( , 0.0056 s−1). In each case, the kinetics of AP phosphorylation were identical within experimental error. Since the KM for ATP for WT Aurora A is about 10 μM (Kelly et al., 2011), all following kinetic reactions were run at 5 mM ATP (20 mM TrisHCl, 200 mM NaCl, 20 mM MgCl2, 3% [vol/vol] glycerol, 1 mM TCEP, pH 7.50) at 25°C. Phosphorylated peptide production was monitored by reverse phase-high performance liquid chromatography (RP-HPLC).
, 0.0056 s−1). In each case, the kinetics of AP phosphorylation were identical within experimental error. Since the KM for ATP for WT Aurora A is about 10 μM (Kelly et al., 2011), all following kinetic reactions were run at 5 mM ATP (20 mM TrisHCl, 200 mM NaCl, 20 mM MgCl2, 3% [vol/vol] glycerol, 1 mM TCEP, pH 7.50) at 25°C. Phosphorylated peptide production was monitored by reverse phase-high performance liquid chromatography (RP-HPLC).
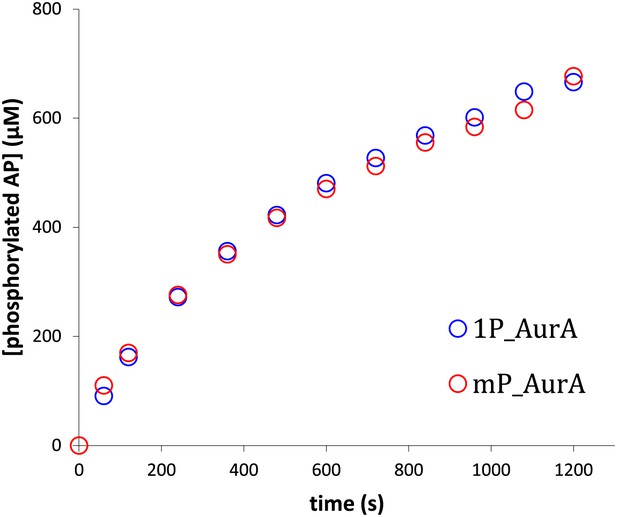
Aurora A exhibits the same activity towards AP whether the protein is phosphorylated on multiple sites or singly phosphorylated on T288.
The rates are 0.93 s−1 and 0.97 s−1, respectively. To obtain singly phosphorylated Aurora A (1P_AurA) , dephosphorylated protein was autophosphorylated in the presence of ATP and a final concentration of 1 μM of this protein was used for the assay described here. Aurora A phosphorylated on multiple sites (mP_AurA) was obtained through classic expression in Escherichia coli cells (see Figure 1A). Reactions were carried out in the presence of 5 mM ATP and 1 mM AP in assay buffer at 25°C.
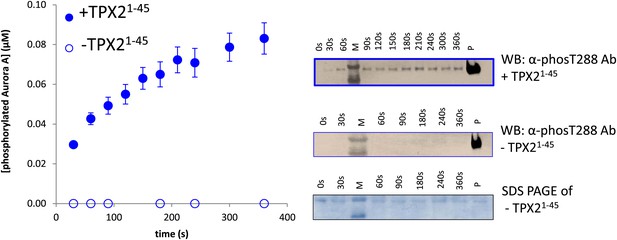
Aurora A kinase autophosphorylation and substrate phosphorylation were simultaneously followed either in the absence or presence of TPX21−45.
In the experiments aimed at measuring activity of AP phosphorylation of dephosphorylated Aurora A (Figure 1B), autophosphorylation can occur during the time-course of the reaction. Therefore autophosphorylation of 1 μM Aurora A in the presence of 1 mM AP and 5 mM ATP and in the absence or presence of 50 μM TPX21−45 was monitored simultaneously with AP phosphorylation. Densitometry analysis (left) of raw Western blot data (right) is shown. To account for Aurora A's dynamic range, time points up to 300 s were diluted 50× and the rest of the time points were diluted 225×. The amount of phosphorylated protein made during the reaction accounts for only 10% of the detected rate acceleration of AP phosphorylation in the presence of TPX2.
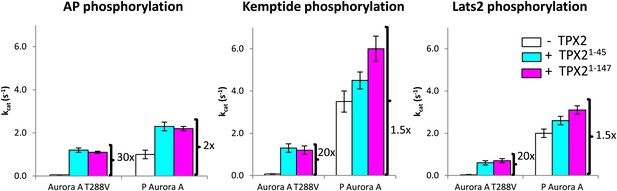
TPX21−45 drastically accelerates the kinetics of the dephosphorylated-like Aurora A species irrespective of the nature of the peptide used.
TPX2 increases the kinetics of the dephosphorylated-protein mimic (T288V mutant) towards the peptides AP (APSSRRTTLCGTL) (left), Kemptide (LRRASLG) (middle), and Lats2373−387 (ATLARRDSLQKPGLE) (right) between 20- and 30-fold. Lats2 is an Aurora A substrate important in centrosome maturation, and Kemptide is a synthetic construct generally used as a substrate of cAMP-dependent protein kinase A (PKA), a protein closely related to Aurora A. Reactions are carried out in the presence of 5 mM ATP and 1 mM peptide in assay buffer at 25°C. A longer TPX2 variant (TPX21−147) was used as control to ensure full capture of TPX2 activity by the shorter variant (TPX21−45). P: phosphorylated.
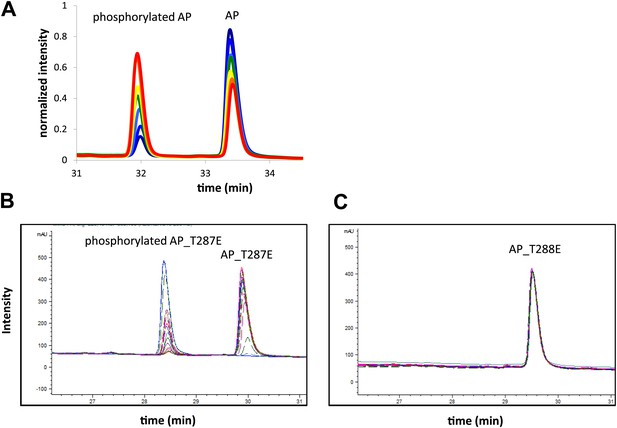
Representative RP-HPLC time traces during AP phosphorylation.
These traces (A) show well resolved non-phosphorylated and phosphorylated peptide peaks at different reaction time points. (B) Screenshot of time traces for AP_T287E (left) and AP_T288E (right) phosphorylation show that Aurora A selectively phosphorylates AP on T288. Assays were carried out in the presence of 1 μM phosphorylated Aurora A, 1 mM AP or 1 mM AP_T287E or AP_T288E, 5 mM ATP, at 25°C in kinase assay buffer (20 mM TrisHCl, 200 mM NaCl, 20 mM MgCl2, 3% glycerol, 1 mM TCEP, pH 7.50).
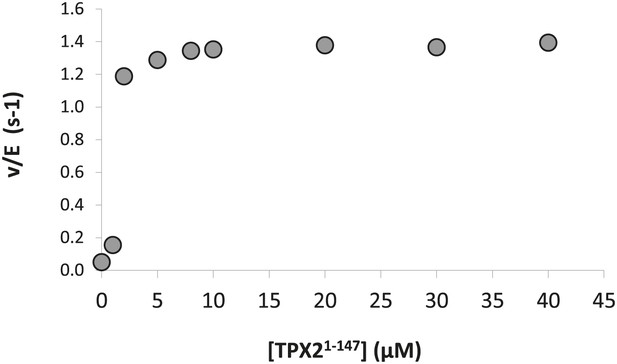
Dose-dependence of the concentration of TPX21−147 on the phosphorylation kinetics of AP by Aurora A T288V.
The calculated KA = 1.0 ± 0.5 μM compared well with the KD = 1.1 ± 0.1 μM obtained from ITC data (Figure 7A). Assays were carried out in the presence of 1 μM Aurora A T288V, 1 mM AP, increasing concentrations of TPX21−147, 5 mM ATP, at 25°C in kinase assay buffer (20 mM TrisHCl, 200 mM NaCl, 20 mM MgCl2, 3% glycerol, 1 mM TCEP, pH 7.50).
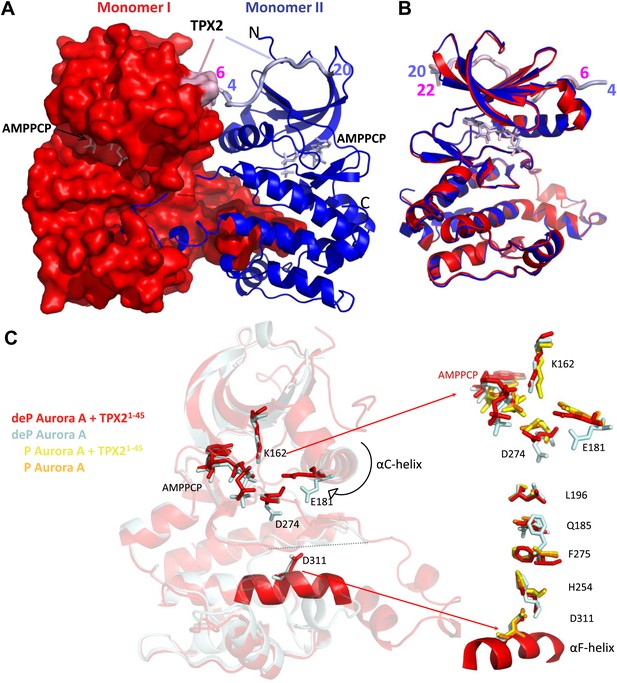
Dephosphorylated Aurora A adopts an active conformation in the presence of TPX21−45.
(A) Dephosphorylated Aurora A + TPX21−45 + AMPPCP and (B) superposition of Aurora A moieties. (C) A detailed view of structural elements that define an active Aurora A kinase: the nucleotide binding region (top inset) and the regulatory spine (bottom inset). Dephosphorylated (deP) Aurora A in the presence of TPX2 (red, PDB ID 4C3P) superposes very well to the phosphorylated (P) Aurora A either in the absence (orange, PDB ID 1OL7) or presence of TPX2 (yellow, PDB ID 1OL5). For comparison, dephosphorylated Aurora A alone (light blue, PDB ID 4C3R) shows the characteristic features of an inactive kinase.
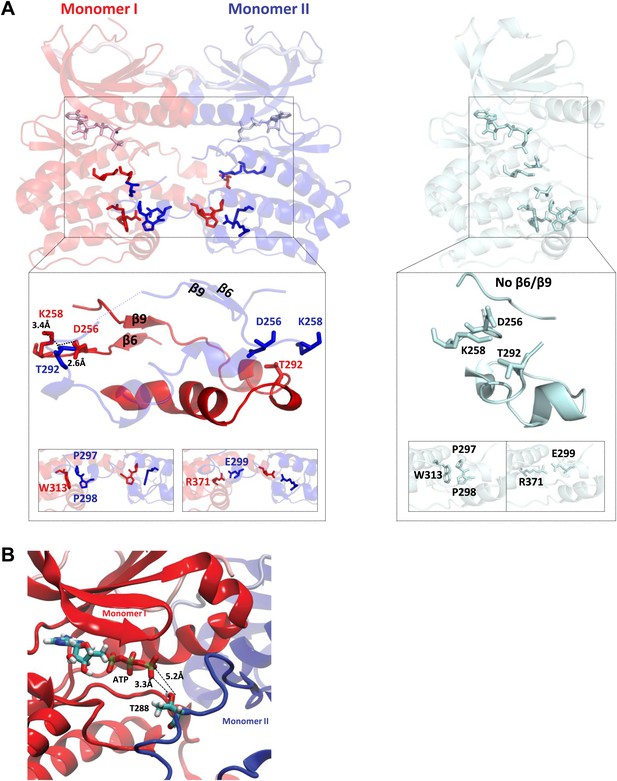
TPX2-bound domain-swapped Aurora A captures an enzyme–substrate complex.
(A) Left: in the presence of TPX21−45, the N-terminal (β6 and β9 sheets) anchor point of the activation loop is present in both monomers whereas the C-terminal H-bond contacts typical for a fully active kinase (between D256/K258 and T292-OH) are only visible for the enzyme monomer in red, which we therefore define as the enzyme molecule. Right: for comparison, in the absence of TPX21−45, the N- and C-terminal anchor points are not present and the protein is in an inactive state. Interactions that further stabilize the swapped dimer (W313-P297/P298 and R371-E299) are shown in the bottom inset, highlighting that these intermolecular interactions (left) are identical to the corresponding intramolecular interactions (right). (B) The loop spanning residues 283–288 in monomer II, for which there was too weak electron density, was remodeled using the software Modeller and biased molecular dynamics. The loop can be arranged by TMD so that the distance between T288 of monomer II and γ-phosphate of AMPPCP of monomer I is compatible with phosphoryl transfer.
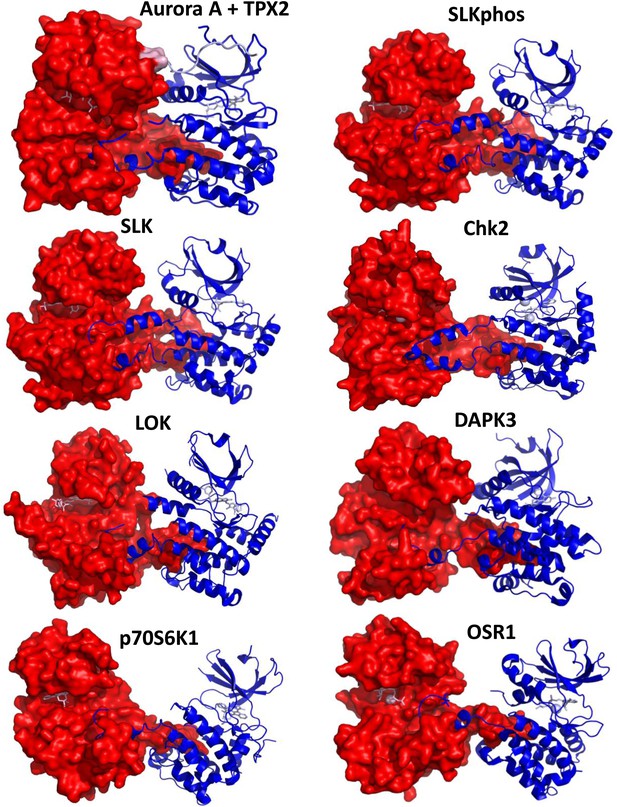
Comparison of the dimeric Aurora A + TPX2 structure with other domain-swapped Ser/Thr kinases.
The activation loop and αEF-helix of one of the monomers nestle between the αF- and αG-helices of the other monomer in the dimer structures. Shown are SLK phosphorylated or non-phosphorylated (PDB IDs 2JFL and 2J51, respectively, both bound to triazole inhibitor DKI), Chk2 (PDB ID 2CN5 bound to ADP), LOK (PDB ID 2 J7T bound to SU11274), DAPK3 (PDB ID 2J90 bound to pyridone 6), p70S6K1 (PDB ID 3A60 bound to staurosporine), and OSR1 (PDB ID 3DAK bound to AMPPNP). One monomer is shown in surface representation and the other in ribbon representation with bound nucleotides shown as sticks (pink). Angles between the monomers and dimeric interface contacts vary significantly.
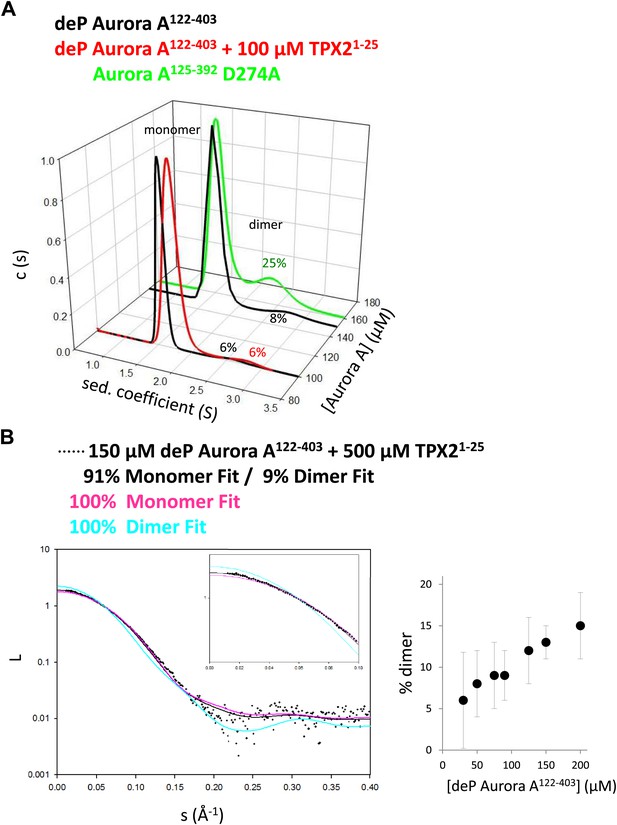
TPX2-bound domain-swapped Aurora A forms a stable dimer in solution.
(A) Sedimentation velocity analytical ultracentrifugation data show discrete peaks for monomer and dimer. TPX21−25 does not increase the percentage of Aurora A dimer in solution. It is unclear why there is an increased dimer concentration for the kinase-dead Aurora A D274A mutant. (B) Small-angle X-ray scattering (SAXS) data show an increase in dimer concentration with increased Aurora A amounts. All data were collected in the presence of 500 μM AMPPCP in kinase assay buffer. deP, dephosphorylated.
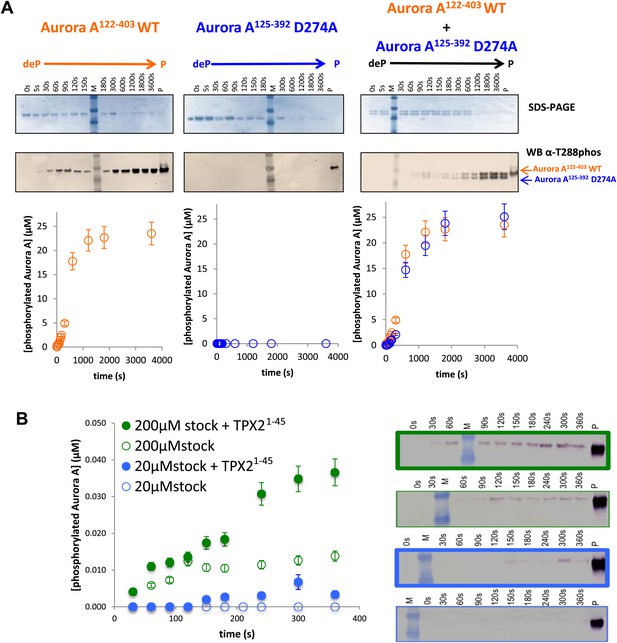
Mechanism of autophosphorylation.
(A) The kinetics of autophosphorylation was monitored by SDS-PAGE and Western blot of 25 μM Aurora A122−403 WT or 25 μM Aurora A125−392 D274A. WT Aurora A can phosphorylate catalytically dead D274A Aurora A intermolecularly. To account for Aurora A's dynamic range, time points up to 300 s were diluted 50-fold and the rest of the time points were diluted 225-fold. (B) Dilution to 1 μM protein from a stock solution of 200 μM Aurora A ± TPX2 shows much faster autophosphorylation kinetics than from a lower concentrated stock solution (20 μM ± TPX2) revealing that autophosphorylation occurs within the long-lived dimer. All experiments were carried out at 25°C in kinase assay buffer in the presence of 5 mM ATP.
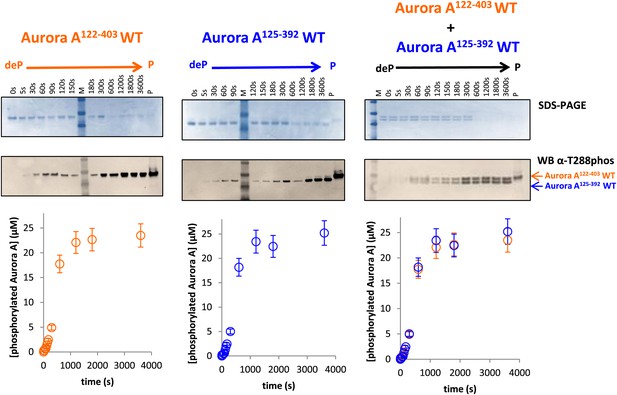
Dephosphorylated Aurora A125−392 can autophosphorylate as efficiently as Aurora A122−403.
SDS-PAGE and Western blot of 25 μM Aurora A122−403 WT or 25 μM Aurora A125−392 show that both proteins can autophosphorylate to similar extents, suggesting that impaired kinetics in Figure 5A are due to the D274A mutation and not the length of the protein construct. To account for Aurora A's dynamic range, time points up to 300 s were diluted 50× and the rest of the time points were diluted 225×. All experiments were carried out as described in Figure 5A. deP: dephosphorylated; P: phosphorylated.
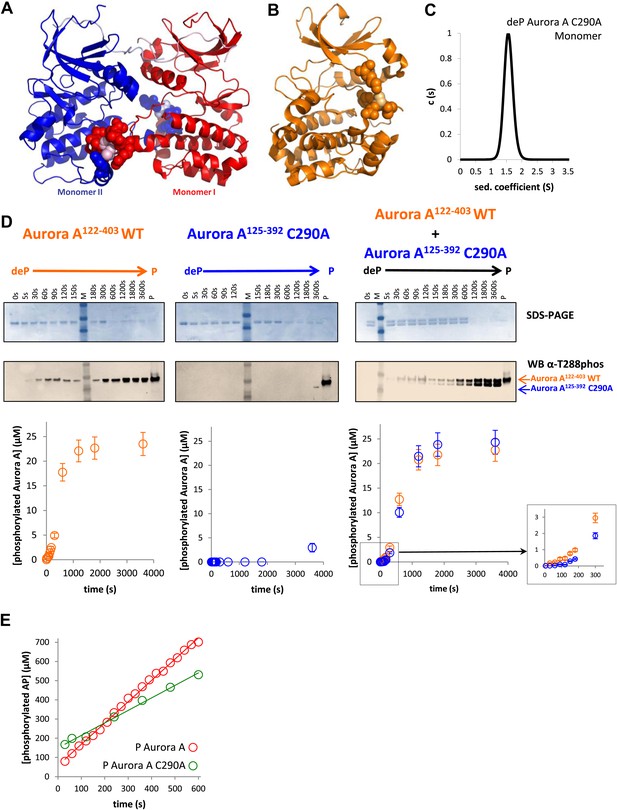
A mutant at the dimer interface (C290A) disrupts the swapped-dimer formation and autophosphorylation without affecting the activity of the phosphorylated Aurora A C290A monomer.
(A) C290 of monomer I (light pink spheres) packs against Y334 of the αG-helix of monomer II. Other residues within a 4.5 Å radius of C290 in monomer I are shown as red spheres. (B) In the monomeric, phosphorylated Aurora A (PDB ID 1OL7), C290 (light orange) does not contact the αG-helix (contact residues within a 4.5 Å radius are shown as orange spheres). (C) Sedimentation velocity analytical ultracentrifugation of 100 μM dephosphorylated (deP) Aurora A C290A + 500 μM AMPPCP in kinase assay buffer shows that this protein is predominately monomeric in solution. (D) The kinetics of autophosphorylation was monitored by SDS-PAGE and Western blot of 25 μM Aurora A122−403 WT or 25 μM Aurora A125−392 C290A as described in Figure 5. C290A mutant has impaired autophosphorylation, but is readily phosphorylated by WT Aurora A. (E) Activity of the phosphorylated (P), monomeric C290A towards AP peptide (0.7 ± 0.1 s−1) is comparable to that of the WT protein (1.0 ± 0.2 s−1).
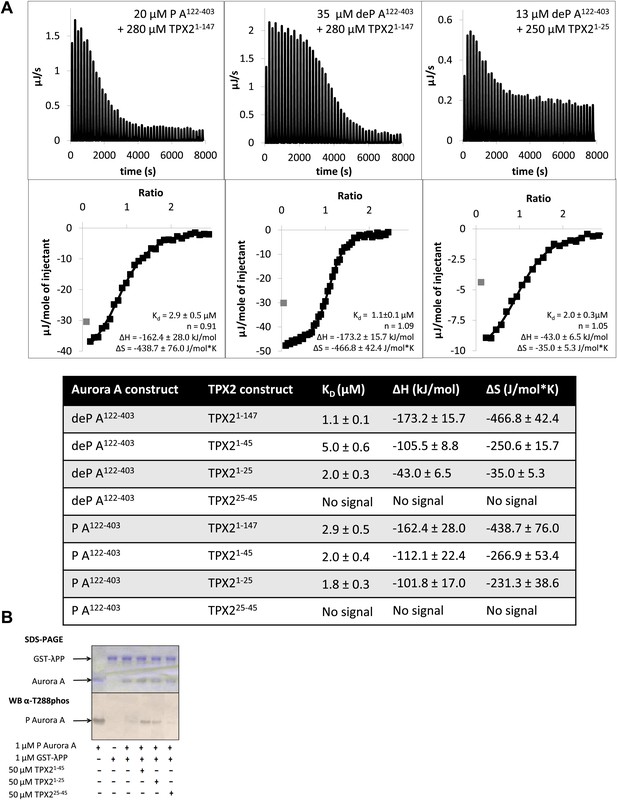
The N-terminal half of TPX21−25 is the minimal region needed for binding to Aurora A.
(A) Isothermal titration calorimetry (ITC) measurements conducted with various TPX2 constructs show that TPX2 binds with similar affinity to either the phosphorylated (P) or the dephosphorylated (deP) Aurora A and that the minimal length required for binding encompasses the first 25 residues of TPX2. (B) At the functional level, TPX21−25 can protect Aurora A from λ protein phosphatase (λPP)-directed dephosphorylation to the same extent as TPX21−45.
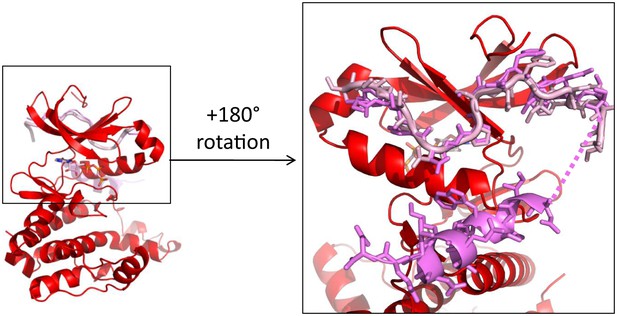
The first 25 amino acids of TPX2 bind similarly to either dephosphorylated or phosphorylated Aurora A.
Superposition of TPX2 from the dephosphorylated Aurora A (red) + TPX2 (light pink) and the phosphorylated Aurora A (not shown) + TPX2 (magenta) (PDB ID 1OL5) shows that the N-terminal half of TPX2 binds similarly to both proteins whereas the C-terminal half of TPX2 only binds to phosphorylated Aurora A that is monomeric in the X-ray structure, but not to the dimeric, dephosphorylated Aurora A. The dotted line represents missing electron density for residues TPX223−29.
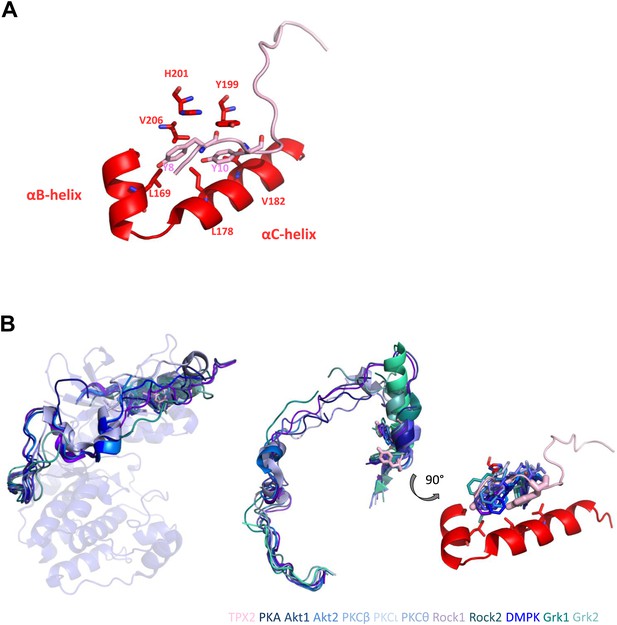
Two conserved tyrosines of TPX2 nestled inside a hydrophobic pocket in Aurora A trigger allosteric activation.
(A) Y8 and Y10 of TPX2 make extensive contacts with residues lining αB- and αC-helices in Aurora A. (B) Superposition of TPX2 bound to Aurora A with the C-terminal tails of several AGC kinases reveals equivalent positioning of Phe of the FxxF hydrophobic motifs in the conserved kinases’ hydrophobic pockets. Right: a zoom into this highly conserved protein/protein interaction motif used for allosteric regulation. We note that while for most AGC kinases this interaction occurs via its own C-terminal tails, in the evolutionarily younger Aurora A kinase, this regulation is mediated by interaction with a second binding partner, TPX2 (Davis et al., 2008).
Tables
Data collection and refinement statistics
| deP Aurora A + AMPPCP | deP Aurora A + AMPPCP + TPX2 | |
|---|---|---|
| Data collection | ||
| Space group | P 61 2 2 | P 21 21 21 |
| Cell dimensions | ||
| a, b, c (Å) | 83.47, 83.47, 172.63 | 49.93, 86.72, 153.55 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 86.3–2.79 (2.87–2.79) | 86.7–2.69 (2.76–2.69) |
| Rmerge | 0.08 (2.08) | 0.26 (3.51) |
| I/σ | 19.8 (1.8) | 8.0 (2.3) |
| Completeness (%) | 100 (100) | 100 (100) |
| Redundancy | 15.7 (16.7) | 6.9 (7.2) |
| Refinement | ||
| Resolution (Å) | 55.4–2.79 (2.87–2.79) | 47.5–2.69 (2.76–2.69) |
| No. reflections | 8459 | 18104 |
| Rwork/Rfree | 0.221/0.306 (0.327/0.451) | 0.201/0.289 (0.284/0.400) |
| No. atoms | ||
| Protein | 2074 | 4574 |
| Ligand/ion | 31 | 67 |
| Water | 0 | 21 |
| B-factors | ||
| Protein | 100.3 | 54.6 |
| Ligand/ion | 109.1 | 62.5 |
| Water | NA | 43.3 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.010 | 0.011 |
| Bond angles (°) | 1.54 | 1.53 |
| PDB ID | 4C3R | 4C3P |
-
Values in parentheses correspond to the highest-resolution shell.
-
deP: dephosphorylated; PBD, Protein Data Bank.





