A molecular model for the role of SYCP3 in meiotic chromosome organisation
Figures
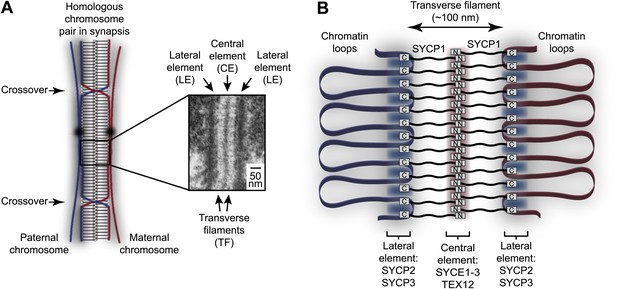
Homologous chromosome synapsis by the synaptonemal complex.
(A) The synaptonemal complex (SC) is a molecular ‘zipper’ that holds together homologous chromosomes during meiotic prophase I, enabling recombination and crossover formation. The SC has a tripartite ultrastructural appearance in which transverse filaments bridge between a midline central element and lateral elements that coat the chromosome axes. The inset electron micrograph is reproduced from Kouznetsova et al. (2011) under the Creative Commons Attribution License. (B) Model for assembly of the mammalian SC from its key components. SYCP1 forms the transverse filaments, with its N- and C-terminal regions located in the central and lateral elements respectively. The central element also contains SYCE1, SYCE2, SYCE3 and TEX12, whilst the lateral element contains SYCP2 and SYCP3.
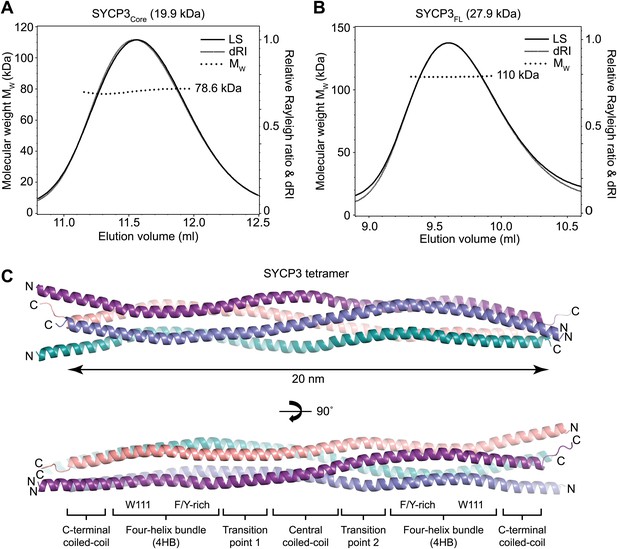
Structure of human SYCP3.
(A and B) SEC-MALS analysis of SYCP3Core and SYCP3FL, in which light scattering (LS) and differential refractive index (dRI) are plotted in conjunction with fitted molecular weights (MW). (A) SYCP3Core has a fitted molecular weight of 78.6 kDa (±0.259%), with polydispersity 1.000 (±0.365%); its theoretical tetramer size is 79.5 kDa. (B) SYCP3FL has a fitted molecular weight of 110 kDa (±0.064%), with polydispersity 1.000 (±0.091%); its theoretical tetramer size is 111 kDa. (C) The crystal structure of SYCP3Core is shown with a 90° rotation around its longitudinal axis; chains A-D are depicted in purple, salmon, teal and blue. The tetramer provides a length of 20 nm between the extremes of its C-terminal coiled-coils (measured at 196.1 and 199.9 Å between Gln220 Cα atoms of chains A and B, and chains C and D, respectively). The structure is made up of a central coiled-coil and two flanking arms. Each arm contains a four-helix bundle (with proximal aromatic-rich and distal Trp111 regions) and a C-terminal coiled-coil at the distal end. The four-helix bundle regions become continuous with the central coiled-coil through transition points that are distinct for each arm.
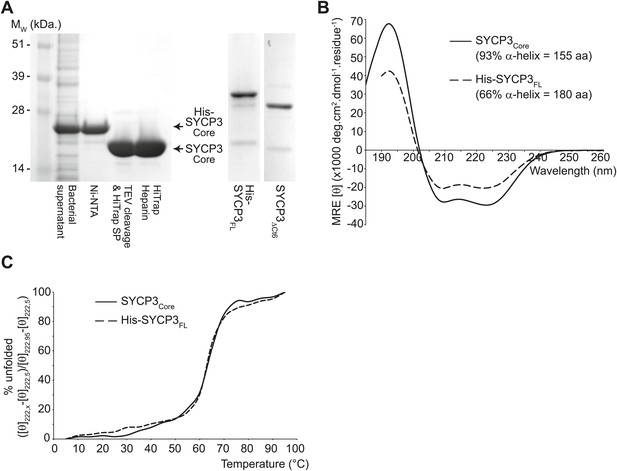
The helical core of human SYCP3 is defined by amino acids 66-230.
(A) Coomassie stained SDS-PAGE summarising the purification of SYCP3Core (amino acids 66–230) by Ni-NTA, TEV-cleavage, HiTrap SP and HiTrap Heparin chromatography (left), and showing purified SYCP3FL (amino acids 1–236) and SYCP3ΔCt6 (amino acids 1–230) samples (right). (B) Far UV CD spectra of SYCP3Core (solid line) and SYCP3FL (dashed line) recorded between 260 nm and 185 nm (truncated at 190 nm for SYCP3FL owing to high background from 450 mM NaF), measured in mean residue ellipticity, MRE ([θ]) (× 1000 deg·cm2·dmol−1·residue−1). Helical content was estimated at 93% (155 amino acids) for SYCP3Core and 66% (180 amino acids) for SYCP3FL through deconvolution using the CDSSTR algorithm, with data fitted at normalised r.m.s. deviation values of 0.008 and 0.011 respectively. (C) CD temperature melts of SYCP3Core (solid line) and SYCP3FL (dashed line), recording the helical signal at 222 nm in MRE ([θ222]) at 5°C intervals between 5 and 95°C. Data are presented as % unfolded, calculated as ([θ]222,x − [θ]222,5)/([θ]222,95 − [θ]222,5).
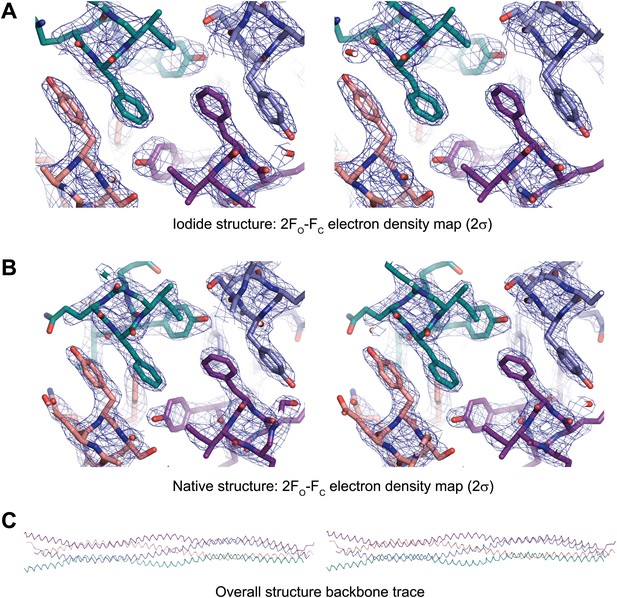
Stereo images of sample electron density and the overall SYCP3 structure.
(A) Sample 2FO-FC electron density map of the SYCP3 iodide structure contoured at 2σ. (B) Sample 2FO-FC electron density map of the SYCP3 native structure contoured at 2σ. (C) Backbone trace of the overall native SYCP3 structure.
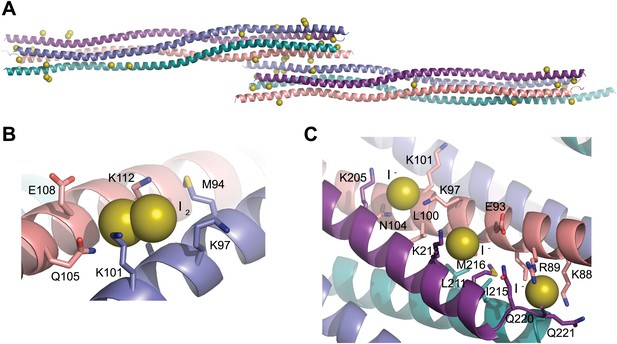
Iodide sites used for SAD phasing.
(A) The SYCP3 iodide derivative crystal contains 43 iodide sites across the two tetramers of the asymmetric unit. (B) 16 sites represent molecular iodine, with one I2 molecule present at the same site on each of the eight protein chains. (C) An example of iodide binding to the surface of the SYCP3 structure; iodide sites were mostly found in patches of lysine residues, with the aliphatic chains providing a hydrophobic core and the functional group giving a positively charged lip to the binding site.
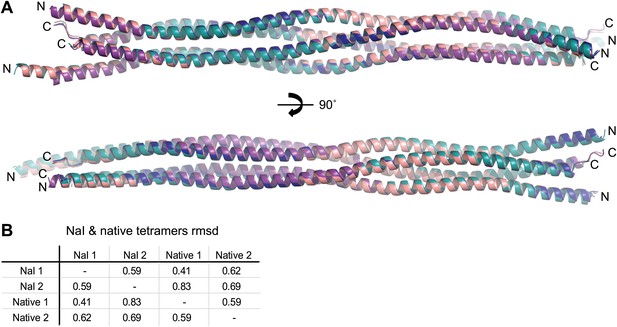
Comparison of SYCP3 tetramers present within native and iodide derivative crystals.
(A) Superposition of the two tetramers present within each of the native and iodide derivative P1 unit cells, coloured as follows: native tetramer 1 (chains A–D)—purple, native tetramer 2 (chains E–H)—salmon, iodide tetramer 1 (chains A–D)—blue, iodide tetramer 2 (chains E–H)—teal. (B) Root mean square deviation (rmsd) values between pairwise superposed native and iodide (NaI) derivative tetramers.
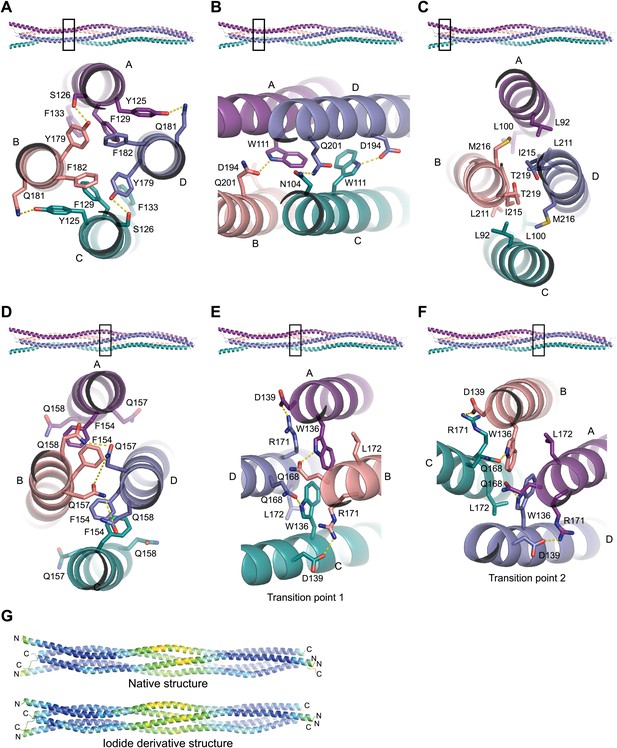
Structural details of the SYCP3 tetramer arm and central coiled-coil regions.
(A–C) The SYCP3 tetramer arm is formed by a bipartite four-helix bundle of a proximal aromatic-rich core and distal W11 region, which becomes continuous with a C-terminal coiled-coil. (A) The aromatic (F/Y)-rich core is assembled through hydrophobic associations of Y125A/C, F129A/C, F133A/C, Y179B/D, and F182B/D, with hydrogen bonds between Y125A/C and Q181B/D, and between Y179B/D and S126A/C. (B) In the distal region, tryptophan residues W111 adopt distinct conformations in chains A and C, undergoing hydrogen bonding with Q201B (W111A) and D194D (W111C). (C) At the distal end of the SYCP3 tetramer, a parallel coiled-coil is formed between the C-terminal ends of chains B and D, involving residues L211B/D, I215B/D, and T219B/D. Flanking chains A and C diverge and are oriented through interactions of L92A/C, L100A/C and M216B/D. (D–F) The central region of SYCP3 is asymmetrical, containing of a parallel coiled-coil flanked by transition points that are distinct between the two tetramer arms. (D) The central parallel coiled-coil is formed between chains B and D, with equivalent chains A and C held apart by steric exclusion. Packing is driven by aromatic interactions between F154 residues, and a network of hydrogen bonds between Q157B/D and Q158B/D (Q157A/C and Q158A/C adopt alternative solvent-exposed conformations). (E) At transition point 1, chains B and D are pulled together for coiled-coil formation through hydrogen bonding between W136A/C and Q168B/D. The interaction is further stabilised by salt bridges between R171B/D and D139C/A. (F) At transition point 2, coiled-coil formation between chains A and C is prevented through an alternative hydrogen bonding pattern in which W111B/D interacts with Q168C/A. Salt bridges between R171A/C and D139D/B are unchanged. (G) The SYCP3 native (top) and iodide derivative (bottom) structures coloured according to their backbone atomic crystallographic B-factors from red (high) to blue (low). Residues of the central region have B-factors of up to four times higher than those of the four-helix bundle regions.
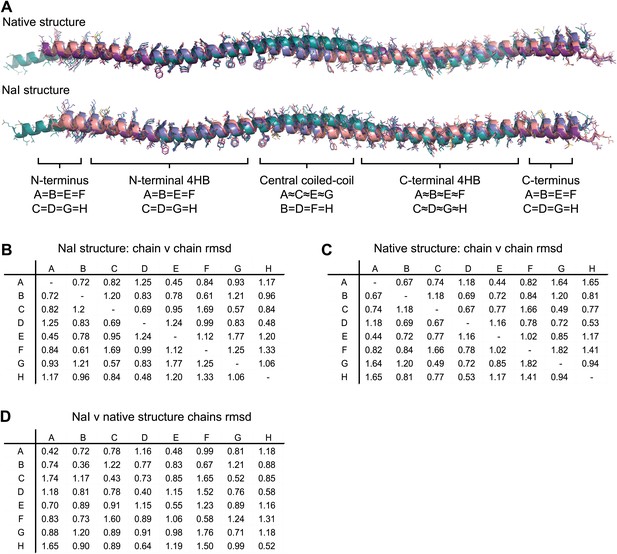
Non-crystallographic symmetry (NCS) between chains of the SYCP3 tetramer.
(A) Superposition of the eight chains that make up the two tetramers of the native and iodide derivative structures. Chains are coloured as follows: A/E–purple, B/F–salmon, C/G–teal and D/H–blue. At the central coiled-coil region, parallel chains are similar (A = C = E = G and B = D = F = H), whilst in the flanking regions anti-parallel chains are similar (A = B = E = F and C = D = G = H). (B–D) Root mean square deviation (rmsd) values between pairwise superposed chains of (B) the iodide derivative structure, (C) the native structure, and (D) between iodide derivative and native structures.
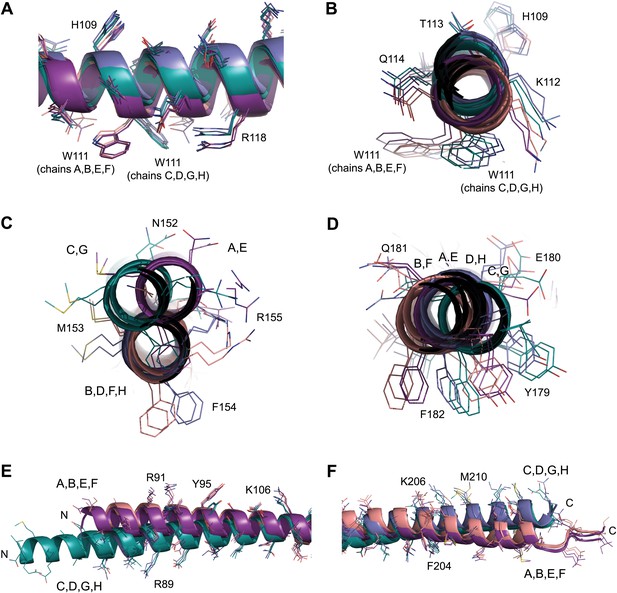
NCS differences between chains of the SYCP3 tetramer.
(A–F) Superposition of the eight chains that make up the two tetramers of the native structure. Chains are coloured as follows: A/E–purple, B/F–salmon, C/G–teal and D/H–blue. (A and B) The W111 residues of the four-helix bundle region adopt distinct conformations between parallel protein chains such that the conformation of chains A, B, E and F differs from that of chains C, D, G and H. (C) In the central coiled-coil region, NCS differences are present between anti-parallel chains. The parallel coiled-coil between chains B and D and F and H holds these chains in the same conformation, whilst flanking chains A and C and E and G show wider conformational freedom. (D) The C-terminal four-helix bundle is the most divergent region, with only approximate similarity between anti-parallel chains A, B, E and F and between C, D, G and H. The (E) N-terminal (F) C-terminal regions of SYCP3 show NCS differences between parallel protein chains.
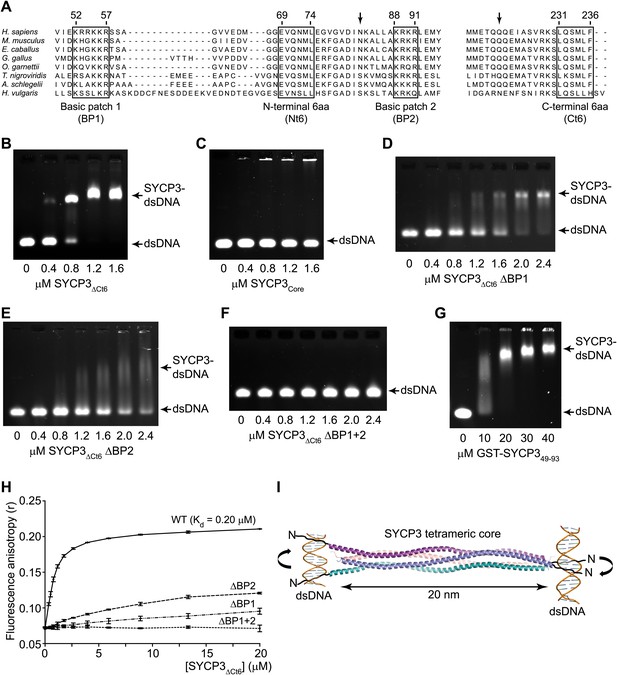
SYCP3 interacts directly with double stranded DNA.
(A) Multiple sequence alignment of the N-terminal (left) and C-terminal (right) regions of SYCP3. The N-terminal region contains two basic patches, BP1 (amino acids 52–57) and BP2 (amino acids 88–91), that flank conserved patch Nt6 (amino acids 69–74). The C-terminus contains conserved patch Ct6 (amino acids 231–236). Arrows indicate N82 and Q221, the N- and C-terminal most amino acids present in all chains of the SYCP3 structure. (B–G) Electrophoretic mobility shift assays in which 187 base pair double stranded DNA (dsDNA) substrates at 18.7 μM (per base pair) were incubated with (B) SYCP3ΔCt6, (C) SYCP3Core, (D) SYCP3ΔCt6 ΔBP1 (mutation of BP1 to alanines), (E) SYCP3ΔCt6 ΔBP2 (mutation of BP2 to alanines), (F) SYCP3ΔCt6 ΔBP1+2 (mutation of BP1 and BP2 to alanines) and (G) GST-SYCP349-93, at concentrations shown. (H) Fluorescence anisotropy analysis in which SYCP3ΔCt6 WT, ΔBP1, ΔBP2 and ΔBP1+2 were incubated with 60 base pair FAM-dsDNA (25 nM per molecule) at concentrations shown. Data points represent the mean and standard deviation (n = 3). The Kd for SYCP3ΔCt6 WT tetramers was determined as 0.20 μM by fitting to a standard curve. (I) Schematic diagram showing how SYCP3 may interact with two DNA molecules. DNA-binding is mediated by BP1 and BP2, which lie at the extreme N-terminus of the SYCP3Core structure. The two ends of the SYCP3 tetramer may bind DNA, leading to two DNA molecules being held apart by 20 nm, with torsional rotation permitted around the longitudinal axis.
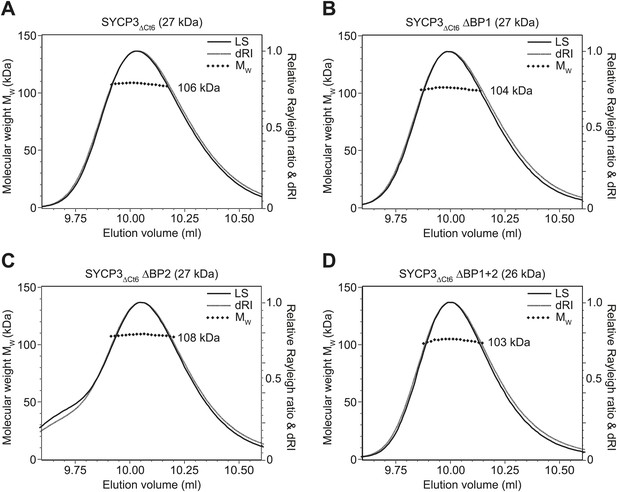
SEC-MALS analysis of SYCP3ΔCt6 and basic patch mutants.
(A–D) SEC-MALS analysis of SYCP3ΔCt6 and basic patch mutants, in which light scattering (LS) and differential refractive index (dRI) are plotted in conjunction with fitted molecular weights (MW). (A) SYCP3ΔCt6 has a fitted molecular weight of 106 kDa (±0.170%), with polydispersity 1.001 (±0.237%); its theoretical tetramer size is 109 kDa. (B) SYCP3ΔCt6 ΔBP1 has a fitted molecular weight of 104 kDa (±0.209%), with polydispersity 1.000 (±0.295%); its theoretical tetramer size is 107 kDa. (C) SYCP3ΔCt6 ΔBP2 has a fitted molecular weight of 108 kDa (±0.145%), with polydispersity 1.000 (±0.205%); its theoretical tetramer size is 107 kDa. (D) SYCP3ΔCt6 ΔBP1+2 has a fitted molecular weight of 103 kDa (±0.231%), with polydispersity 1.000 (±0.327%); its theoretical tetramer size is 106 kDa.
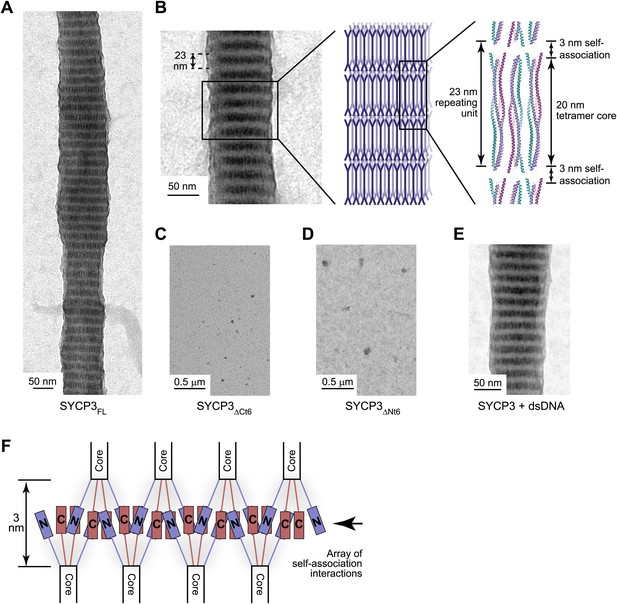
SYCP3 undergoes self-assembly into regular filamentous structures.
(A) Transmission electron micrograph of SYCP3FL, loaded onto an EM grid at 1 mg/ml (32 μM) in buffer containing 250 mM KCl, with negative staining performed using uranyl acetate. (B) SYCP3 fibres vary in length and width but show a constant pattern of light and dark striations, with a periodicity of 23 nm (mean = 23.3 nm, standard deviation = 0.95 nm). These striations may be explained by SYCP3 tetramers lying along the longitudinal axis, with the 20 nm rigid rod providing the bulk of the 23 nm spacing. (C and D) Transmission electron micrographs of 1 mg/ml (C) SYCP3ΔCt6 and (D) SYCP3ΔNt6. (E) Transmission electron micrograph of 1 mg/ml SYCP3FL incubated with 350 base pair double-stranded DNA (dsDNA) at 190 μM (per base pair). (F) The N- and C-terminal regions of SYCP3 are implicated in self-assembly. They are predicted to interact in an interlaced fashion within the remaining 3 nm space, creating arrays of self-association sites within discrete layers that define three-dimensional lattice assembly of SYCP3 fibres.
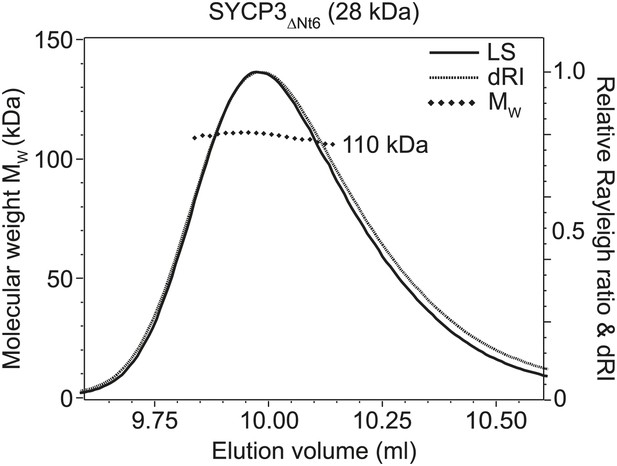
SEC-MALS analysis of SYCP3ΔNt6.
SEC-MALS analysis in which light scattering (LS) and differential refractive index (dRI) are plotted in conjunction with fitted molecular weights (MW). SYCP3ΔNt6 has a fitted molecular weight of 110 kDa (±0.133%), with polydispersity 1.000 (±0.188%); its theoretical tetramer size is 110 kDa.
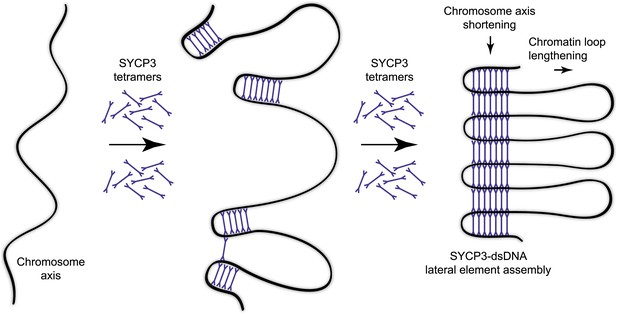
Model for organisation of the chromosome axis by SYCP3 assembly.
Each SYCP3 tetramer contains two DNA-binding regions, separated by a distance of 20 nm owing to the central rigid rod-like structure. Upon binding to the chromosome axis, SYCP3 tetramers may pinch off portions of the axis such that short stretches of chromosomal DNA are looped back on themselves with a separation of 20 nm. The loading of further SYCP3 tetramers may bridge between the initial pinched-off portions, creating a continuous structure that extends along the chromosome axis. Thus, the final assembly consists of a three-dimensional lattice of SYCP3 tetramers that organise the chromosome axis in a concertina-like manner such that the length of the axis is shortened and the chromatin loops (that will flank the SC) are lengthened. For clarity, other meiotic factors that are known to perform important functions in the organisation of the chromosome axis, such as SYCP2, cohesin and condensin, are not depicted.
Tables
Data collection, phasing and refinement statistics
| SYCP3—Iodide SAD | SYCP3—Native MR | |
|---|---|---|
| Data collection | ||
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 49.18, 90.30, 104.22 | 49.14, 92.38, 103.40 |
| α, β, γ (°) | 108.25, 101.20, 102.75 | 66.53, 82.32, 76.53 |
| Resolution (Å) | 47.45–2.41 (2.47–2.41) | 47.36–2.24 (2.29–2.24) |
| Rmerge | 0.064 (0.351) | 0.151 (1.094) |
| I/σI | 15.0 (2.8) | 11.0 (2.5) |
| Completeness (%) | 94.4 (81.2) | 86.7 (38.9) |
| Redundancy | 3.8 (3.0) | 9.7 (8.1) |
| Refinement | ||
| Resolution (Å) | 47.45–2.41 (2.49–2.41) | 47.36–2.24 (2.30–2.24) |
| No. reflections | 106175 (7116) | 66411 (1883) |
| Rwork/Rfree | 0.207/0.229 (0.324/0.354) | 0.195/0.226 (0.245/0.270) |
| No. atoms | 10132 | 10412 |
| Protein | 9751 | 9885 |
| Ligand/ion | 43 | 0 |
| Water | 338 | 527 |
| B-factors | 56.5 | 61.3 |
| Protein | 56.8 | 61.8 |
| Ligand/ion | 72.0 | – |
| Water | 48.3 | 53.6 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.010 |
| Bond angles (°) | 0.870 | 1.030 |
-
One crystal was used for iodide SAD structure solution; one native crystal was used (with data merged from 3 datasets) for molecular replacement. Values in parentheses are for highest-resolution shell.





