Nuclear envelope protein MAN1 regulates clock through BMAL1
Figures
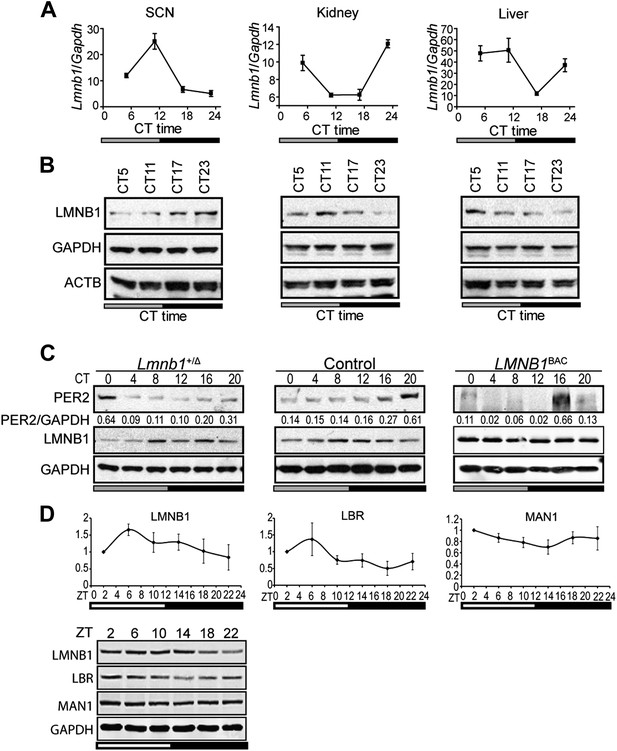
Lamin b1 regulates the circadian clock.
Expression levels of lamin b1 from SCN, kidney, and liver extracts in C57BL/6J mice (A and B). (A) mRNA levels of Lmnb1 and Gapdh were assayed at indicated circadian times (CT, n = 4). (B) Representative immunoblots show the levels of LMNB1, GAPDH, and β-ACTIN. (C) Representative immunoblots show PER2 (with intensity values indicated at the bottom) and LMNB1 abundance in Lmnb1+/Δ, wild-type and LMNB1BAC liver extracts. (D) Expression patterns of LMNB1, LBR, and MAN1 in C57BL/6 mouse livers at indicated Zeitgeber times (ZT) (n = 3). Quantifications (top panels) of Western blots (bottom panel) were obtained by using GAPDH as a loading control. Data represent means ± SD.
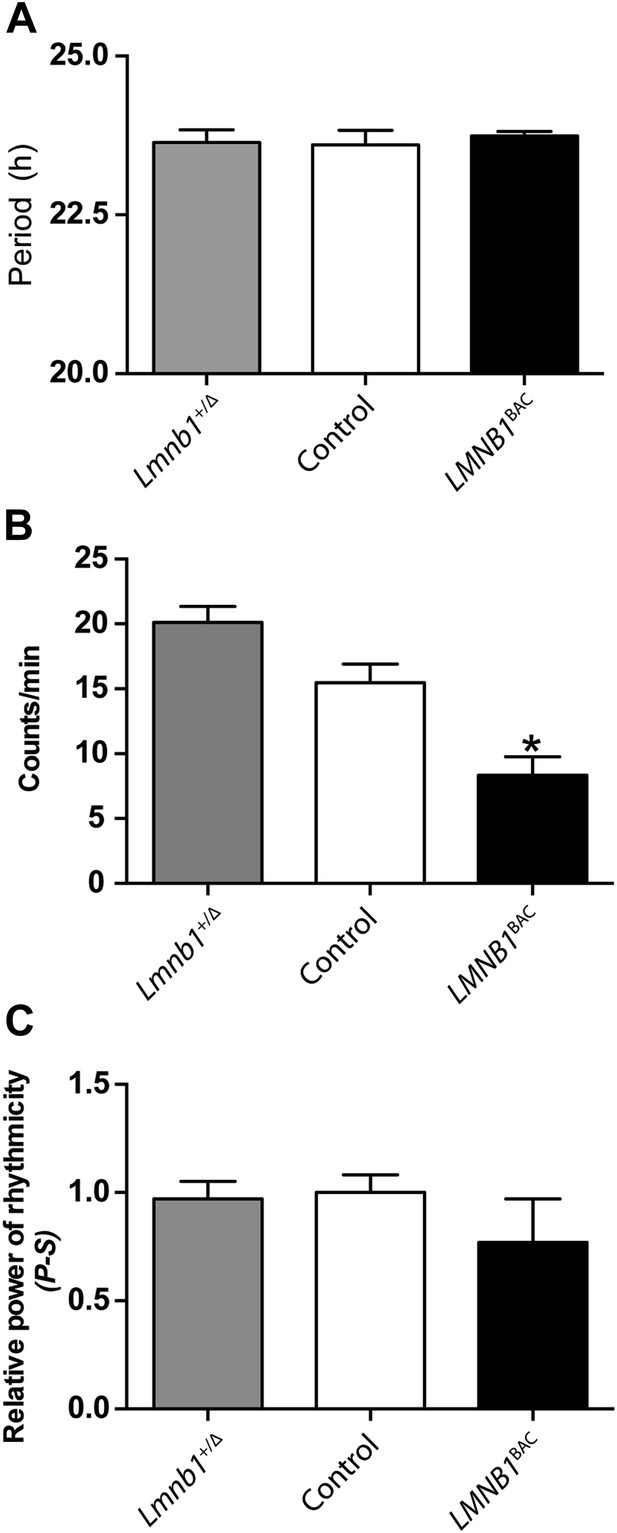
Mice with altered LMNB1 levels do not exhibit altered behavioral rhythms.
The period (A), activity level (B), and amplitude (C) of wheel-running rhythms under DD condition (n = 9–27, Student's t test, *p < 0.05). Error bars represent SEM.
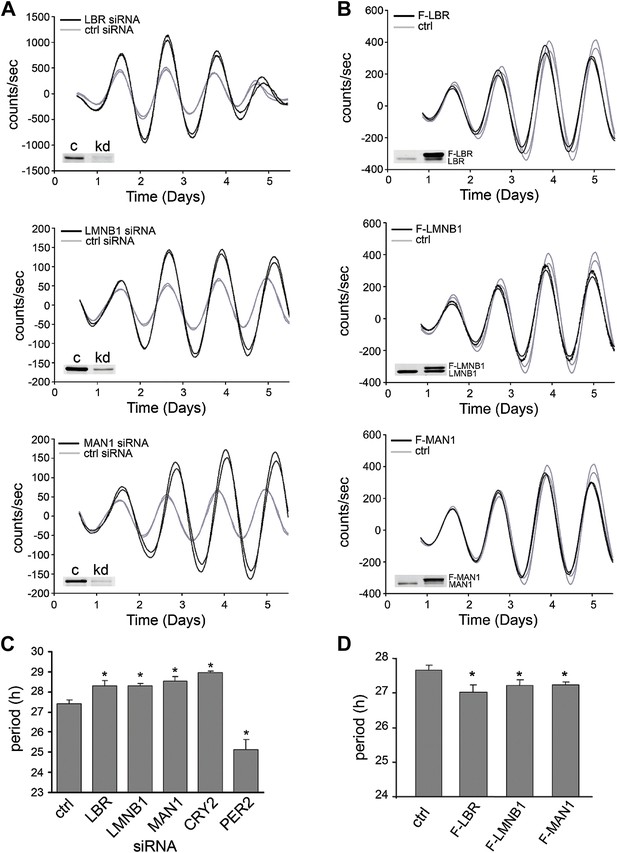
LBR, LMNB1, and MAN1 are necessary for normal circadian rhythms.
Two representative traces of real-time bioluminescence analyses are shown for each, and Western blot verification of down-regulation or over-expression is demonstrated in the inset images. (A) Period was lengthened when LBR, LMNB1, or MAN1 was knocked down. (B) Over-expression of FLAG-tagged LBR (F-LBR), LMNB1 (F-LMNB1), or MAN1 (F-MAN1) shortened period compared to cells transfected with empty vector (ctrl). (C and D) Summary of period in (A and B). CRYPTOCRHOME2 (CRY2) and PER2 siRNA knockdowns served as positive controls. Data represent means ± SD.
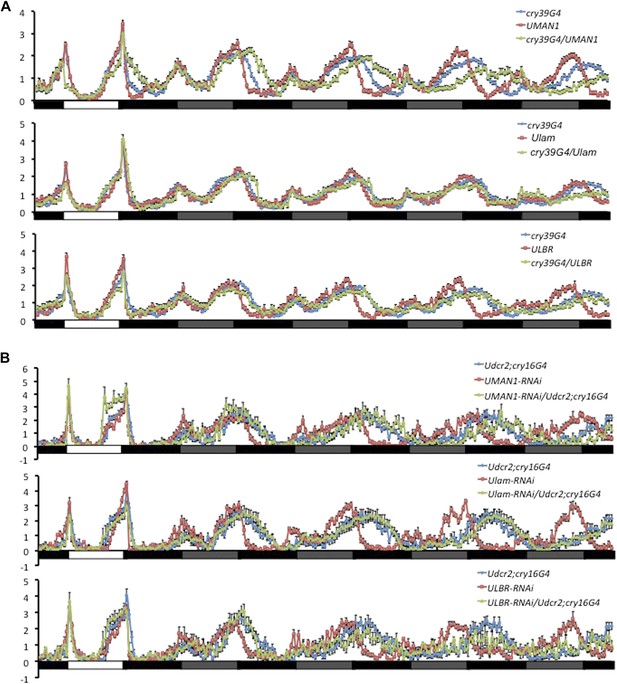
Over-expressing or knocking down nuclear envelope components alters circadian rhythms in flies.
(A) Normalized locomotor activity profiles of flies over-expressing MAN1, Lam, or LBR during LD for 1 day followed by 4 days of DD. (B) Normalized locomotor activity profiles of flies with MAN1, Lam, or LBR knocked down by RNAi. White box indicates light period, black box indicates dark period, and gray box indicates subjective light period. Error bars represent SEM (n = 13–76).
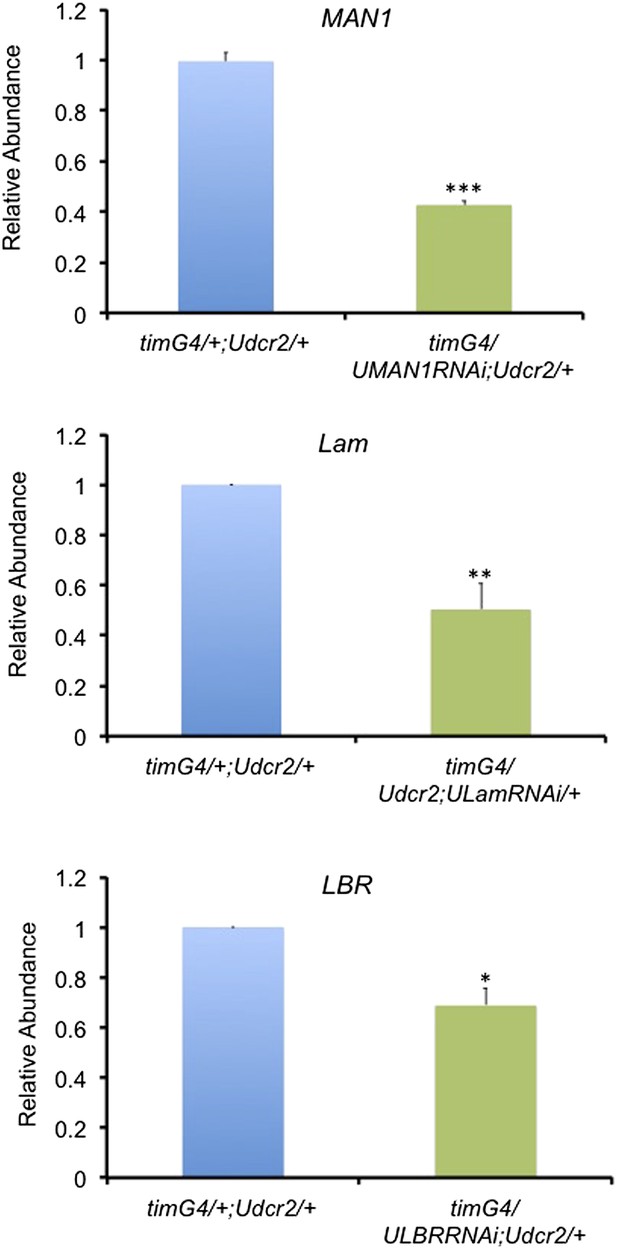
The mRNA levels of nuclear envelope genes are reduced in the corresponding knockdown flies.
Plots of relative mRNA abundance for MAN1, Lam, and LBR from whole head extracts of timGAL4/+;UASdicer2/+, timGAL4/UASMAN1RNAi;UASdicer2/+, timGAL4/+;UASdicer2/UASLamRNAi and timGAL4/UASLBRRNAi;UASdicer2/+ flies determined by qRT-PCR. Error bars represent SEM (n = 3–6). Significant differences indicated by asterisks (Student's t test, *p < 0.05, **p < 0.01, ***p < 0.001). The value of timGAL4/+;UASdicer2/+ for one experiment was set to 1.
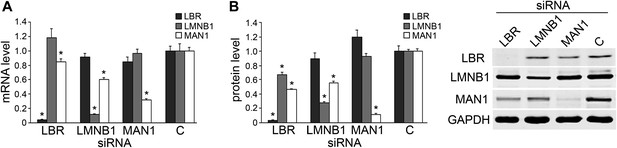
Knocking down LBR/LMNB1 reduces MAN1 mRNA and protein levels but not vice versa.
Assessing mRNA (A) and protein (B) levels of LBR, LMNB1, and MAN1 while knocking them down one at a time in U2OS cells via RNAi. (A) mRNA levels of LBR, LMNB1, and MAN1 in each of the three knockdown conditions were quantified using qRT-PCR (n = 14, *p < 0.05). (B) MAN1 was significantly down-regulated when LBR or LMNB1 was knocked down (n = 14 *p < 0.001). The error bars represent SEM (left panel). Representative immunoblots show the protein levels of LBR, LMNB1 and MAN1 (right panel).
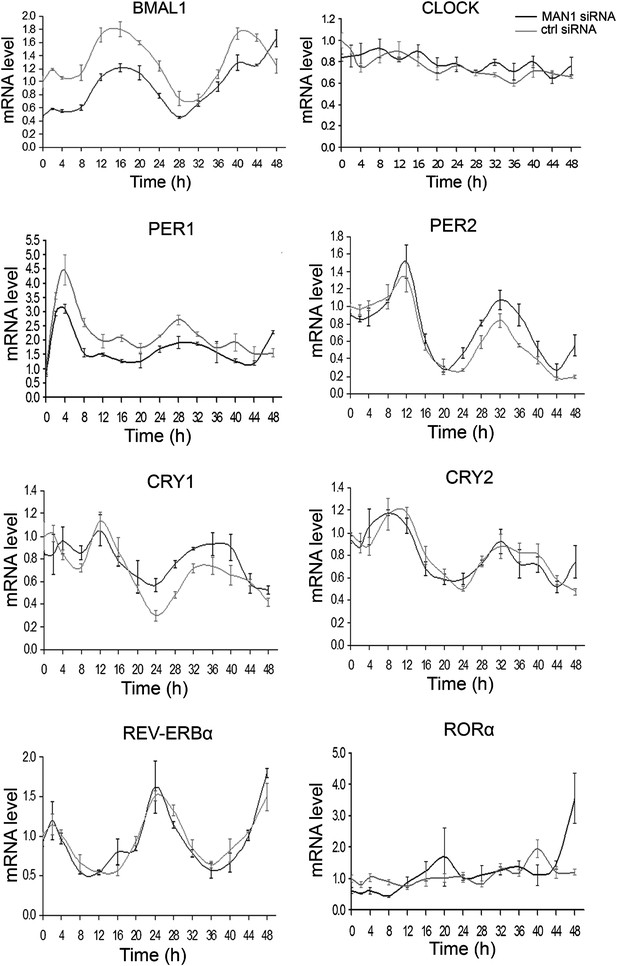
Knocking down MAN1 reduces the levels of BMAL1 mRNA.
Each graph shows cells transfected with MAN1 siRNA vs ctrl siRNA. Time 0 indicates the moment that U2OS cells were treated with dexamethasone (100 nM). Data are presented as means ± SEM, n = 3.
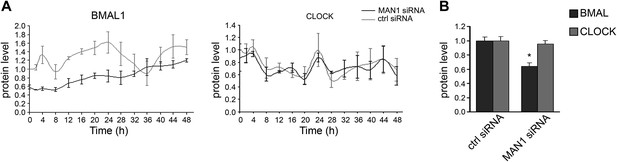
Knocking down MAN1 reduces BMAL1 protein levels.
(A) Time course evaluations of U2OS cells transfected with MAN1 siRNA showed robust reduction of BMAL1 protein levels compared to ctrl siRNA. CLOCK levels were unaffected by MAN1 abundance (n = 3). (B) The quantification results of (A) were graphed (n = 14). Error bars indicate SEM.
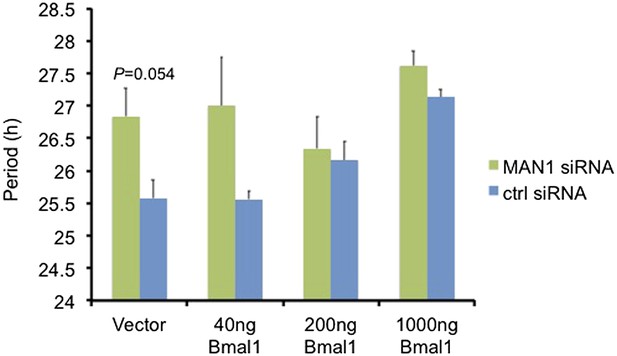
Over-expressing Bmal1 suppresses the period lengthening effect of MAN1 knockdown.
Periods of Bmal1-Luc U2OS cells transfected with MAN1 siRNA or control (ctrl) siRNA together with either empty vector or varying amounts of Bmal1. Error bars represent SEM (n = 2–4, Student's t test).
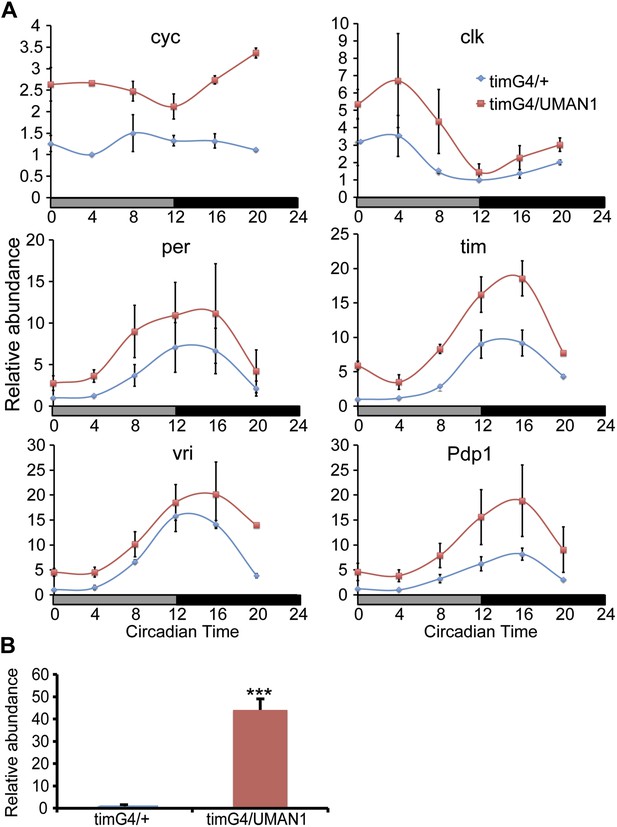
MAN1 increases cyc mRNA levels.
(A) Plots of relative mRNA abundance vs CT for clock genes from whole head extracts of timGAL4/+ and timGAL4/UASMAN1 flies during the first day of DD determined by qRT-PCR (n = 2). Gray box indicates subjective light period and black box indicates dark period. Significant effect of genotypes (Two-way ANOVA) were found for cyc (p = 0.0278) and tim (p = 0.0161). For each time series, the value of the lowest time point was set to 1. (B) Plot of relative mRNA abundance for MAN1 from whole head extracts of timGAL4/+ and timGAL4/UASMAN1 flies determined by qRT-PCR (n = 6, Student's t test, ***p < 0.001). The value of timGAL4/+ in one experiment was set to 1. Error bars represent SEM.
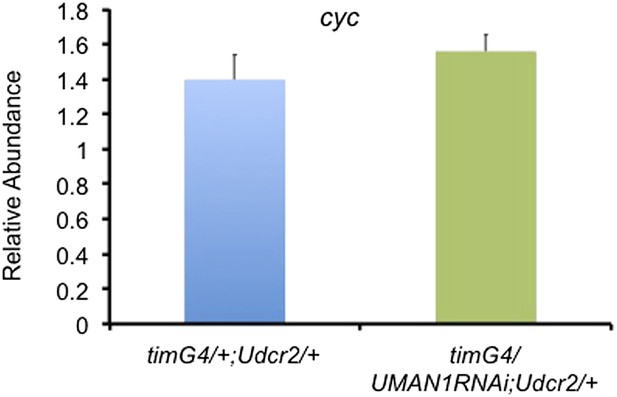
cyc transcript level is not altered in MAN1 knock-down flies.
Plot of relative mRNA abundance for cyc from whole head extracts of timGAL4/+;UASdicer2/+ and timGAL4/UASMAN1RNAi;UASdicer2/+ flies determined by qRT-PCR. Error bars represent SEM (n = 6). The value of timGAL4/+;UASdicer2/+ for one experiment was set to 1.
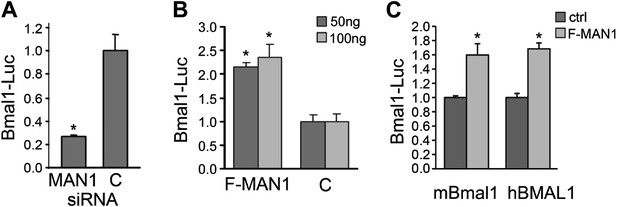
MAN1 promotes BMAL1 transcriptional activity.
(A) Reduction of MAN1 transcripts (13 nM siRNA) reduced Bmal1 promoter activity (n = 3, *p < 0.001). (B) Over-expression of FLAG-tagged MAN1 (F-MAN1) enhanced Bmal1-Luc activity (n = 3, *p < 0.001). (C) Over-expression of FLAG-tagged MAN1 (F-MAN1) enhanced luciferase activities driven by mBmal1 promoter (530 bp) or hBMAL1 promoter (3.4 kb) (n = 3, *p < 0.05). Error bars represent SD.
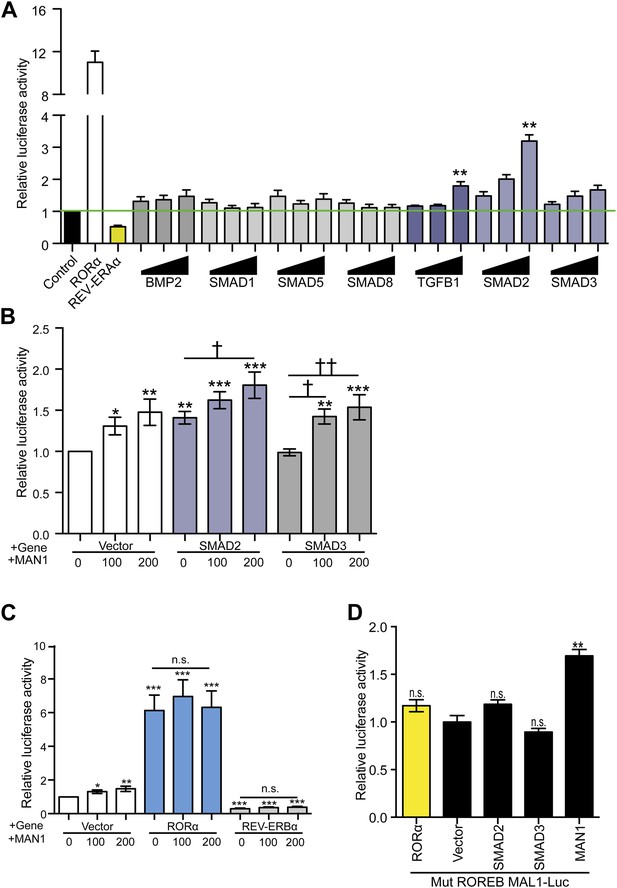
MAN1 and SMAD2 enhance BMAL1 transcription.
(A–D) Luciferase reporter activities in transfected HEK293 cells. Cells transfected with indicated constructs in the presence of the 3.4 kb hBMAL1 promoter for 48 hr and relative luciferase activities were measured in extracts and normalized to Renilla luciferase activities. Relative luciferase activities were shown on the y-axis. Values are means ± SEM, n = 3, **p < 0.01 compared to control. †p < 0.05, ††p < 0.01 compared to MAN1 0, one-way ANOVA with Newman–Keuls test.
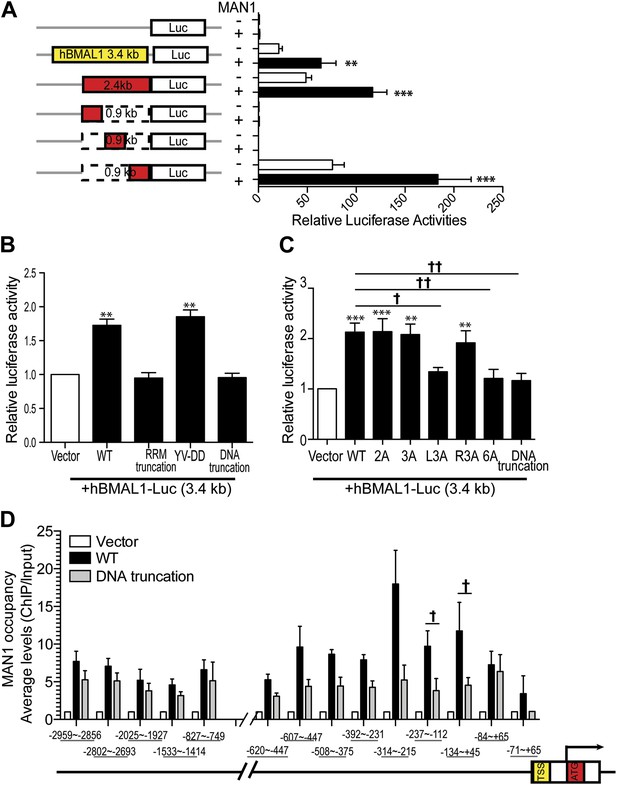
MAN1 binds to the BMAL1 promoter to enhance its transcription.
(A) Luciferase activities of deleted hBMAL1-promoter constructs in the absence or presence of MAN1 expression vectors. n = 3, Student's t test, **p < 0.01, ***p < 0.001. (B and C) Luciferase activities of the 3.4 Kb hBMAL1-Luc in the presence of MAN1 constructs as indicated. n = 3, **p < 0.01, ***p < 0.001 compared to control; †p < 0.05; ††p < 0.001 compared to WT MAN1. (D) ChIP analysis of MAN1 (WT or DNA binding truncation) for 14 segments of hBMAL1 promoter region. Data represent pull-down relative to input. n = 6, †p < 0.05, compared to WT MAN1. One-way ANOVA with Newman–Keuls test. All data are presented as ratio of means ± SEM.
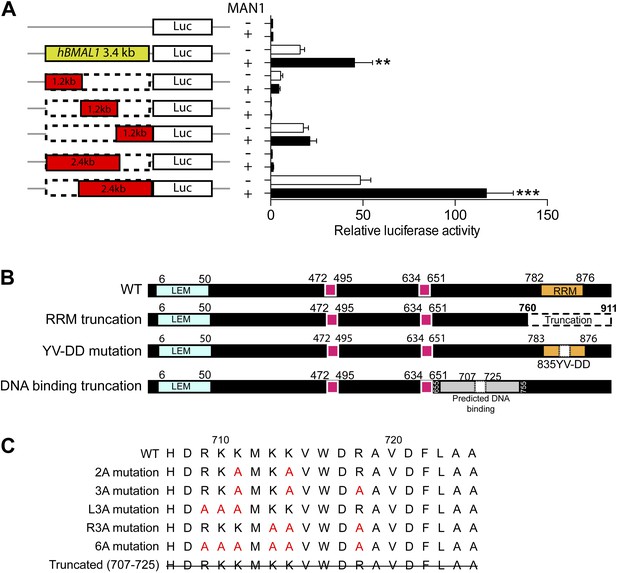
Domain-specific interactions between MAN1 and the hBMAL1 promoter.
(A) Schematic representation of the indicated constructs generated from the 3.4 kb hBMAL1 promoter and cloned into the luciferase reporter vector pGL3. Histogram of luciferase activity in HEK293 cells transfected with the deleted hBMAL1 promoter constructs in the absence or presence of MAN1 expression vectors. Cells transfected for 48 hr and relative luciferase activities measured in extracts and normalized to Renilla luciferase activities. Activities (relative luciferase activity) are shown on the y-axis. Values are means ± SEM, n = 3, Student's t test, **p < 0.01, ***p < 0.001 compared to control. (B) Schematic representation of the deletion constructs generated from the MAN1 expressing construct. (C) Sequences of constructs with point mutations generated from the MAN1 expressing construct.
Tables
Over-expressing NE genes alters the behavioral period in flies
| Genotype | Period (hr) | Power | Rhythmic% | N |
|---|---|---|---|---|
| UASMAN1/+ | 23.8 ± 0.0 | 90 ± 5 | 96 | 76 |
| cryGAL4-39/UASMAN1 | 26.4 ± 0.3*,† | 31 ± 6*,† | 64 | 28 |
| UASMAN1/+; cryGAL4-16/+ | 26.8 ± 0.2*,† | 40 ± 7*,† | 66 | 32 |
| UASLam/+ | 23.9 ± 0.0 | 87 ± 6 | 93 | 55 |
| cryGAL4-39/UASLam | 23.8 ± 0.2† | 33 ± 7*,† | 73 | 22 |
| UASLMNB1/+; cryGAL4-16/+ | 24.4 ± 0.2† | 5 ± 2*,† | 26 | 31 |
| UASLBR/+ | 23.7 ± 0.0 | 59 ± 5 | 79 | 67 |
| cryGAL4-39/UASLBR | 25.6 ± 0.1*,† | 44 ± 6† | 79 | 33 |
| UASLBR/+; cryGAL4-16/+ | N/A | 1 ± 0*,† | 0 | 32 |
| cryGAL4-39/+ | 24.8 ± 0.1 | 86 ± 5 | 93 | 57 |
| cryGAL4-16/+ | 25.6 ± 0.1 | 86 ± 5 | 94 | 64 |
-
*
One-way ANOVA compared to UAS control lines, p < 0.001.
-
†
One-way ANOVA compared to GAL4 control lines, p < 0.001.
Knocking-down NE genes lengthens the behavioral period in flies
| Genotype | Period (hr) | Power | %Rhythmic | N |
|---|---|---|---|---|
| UASMAN1RNAi | 24.4 ± 0.1 | 110 ± 10 | 100 | 14 |
| UASMAN1RNAi;cryGAL4-39/+; UASdcr2/+ | 26.3 ± 0.3*,† | 29 ± 6*,† | 80 | 15 |
| UASMAN1RNAi; UASdcr2/+;cryGAL4-16/+ | 27.9 ± 0.4*,† | 39 ± 8*,† | 69 | 16 |
| UASLamRNAi/+ | 23.6 ± 0.1 | 129 ± 12 | 100 | 16 |
| cryGAL4-39/+; UASLamRNAi/UASdcr2 | 25.1 ± 0.4* | 26 ± 17*,† | 50 | 14 |
| UASLamRNAi/UASdcr2; cryGAL4-16/+ | 26.8 ± 0.1*,† | 126 ± 17 | 100 | 13 |
| UASLBRRNAi/+ | 23.5 ± 0.0 | 112 ± 14 | 100 | 15 |
| cryGAL4-39/UASLBRRNAi; UASdcr2/+ | 24.9 ± 0.1 | 61 ± 12* | 80 | 15 |
| UASLBRRNAi/UASdcr2; cryGAL4-16/+ | 24.6 ± 1.7 | 12 ± 4*,† | 44 | 16 |
| cryGAL4-39/+; UASdcr2/+ | 24.9 ± 0.1 | 69 ± 12 | 88 | 16 |
| UASdcr2/+;cryGAL4-16/+ | 26.1 ± 0.1 | 87 ± 10 | 94 | 16 |
-
*
One-way ANOVA compared to UASRNAi control lines, p < 0.05.
-
†
One-way ANOVA compared to control lines with GAL4 and UASdcr2, p < 0.05.
-
dicer2 (dcr2) is co-expressed to enhance the effects of RNAi.
Additional files
-
Supplementary file 1
(A) Sequences of siRNAs used in the study. (B) qRT-PCR primer sequences for human clock genes.
- https://doi.org/10.7554/eLife.02981.020





