Induction of homologous recombination between sequence repeats by the activation induced cytidine deaminase (AID) protein
Figures
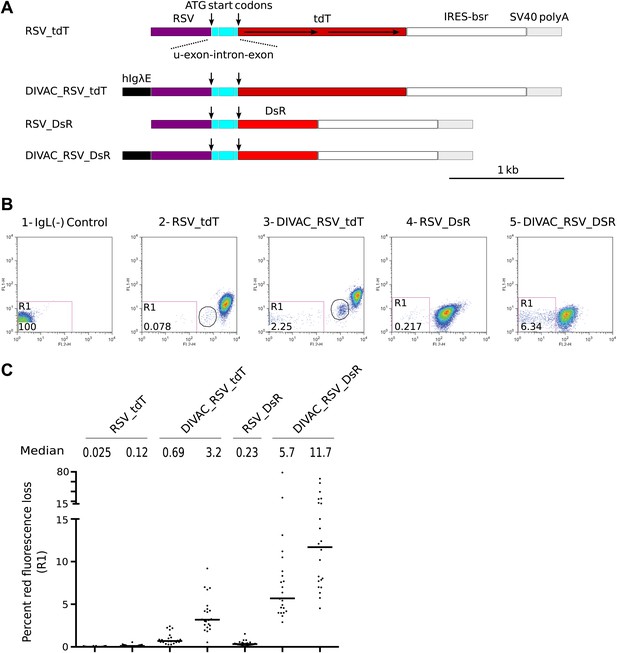
FACS analysis of DT40 transfectants expressing red fluorescence reporter constructs.
(A) Diagrams of the constructs. Sequences important for the behavior of the construct are labeled and color coded: hIgλE—human Igλ enhancer; RSV—rous sarcoma virus promoter; u-exon-intron-exon—upstream splice cassette; tdT and DsR–Tomato and DsRed open reading frames, respectively; IRES-bsr–internal ribosome entry site followed by the blasticidin resistance open reading frame; SV40 polyA—SV40 polyadenylation signal. The names of the constructs indicate the presence of DIVACs, the promoter, the fluorescence genes and sequence repeats. The tandem repeats of tdT are marked by lines with arrows. (B) Two color FACS dot plots of the non-transfected IgL(−) cell line and representative subclones derived from primary transfectants. The levels of green and red fluorescence are plotted according to the x-axis (FL1) and y-axis (FL2), respectively. The number of the plot and the name of the transfected construct are indicated above. Gates and the percentage of gated cells are indicated within the plots. Populations of intermediate red fluorescence are circled. (C) Graphs showing the percentages of gated cells for all subclones of each independent transfectant. The median percentage of gated cells is indicated by the bar and numerically displayed above the graph for each transfectant.
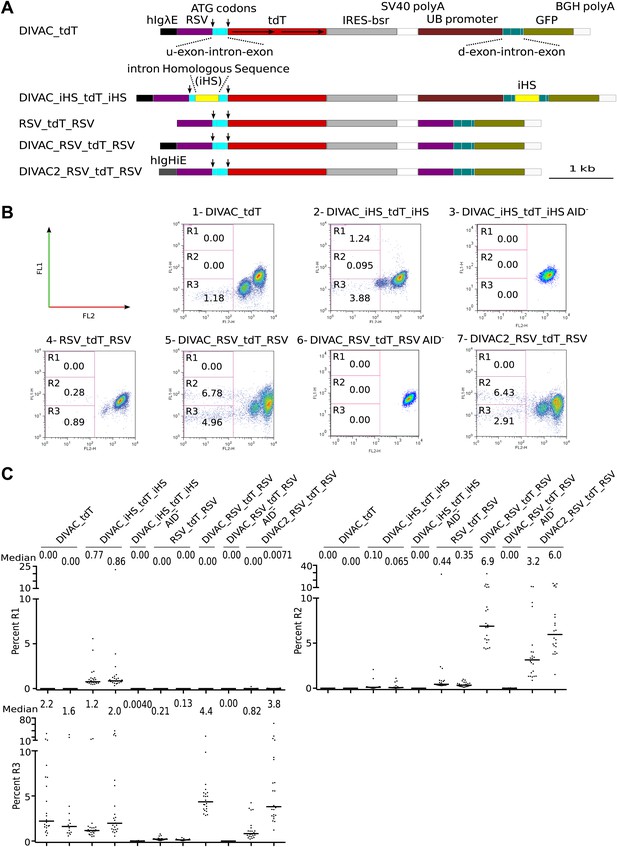
FACS analysis of DT40 transfectants expressing dual tdT/GFP fluorescence reporter constructs.
(A) Diagrams of the constructs. Sequences important for the behavior of the construct are labeled and color coded as in Figure 1 and explained in the following: hIgHiE—human Ig heavy chain intron enhancer; UB promoter—human Ubiquitin C promoter; d-exon-intron-exon—downstream splice cassette; GFP—GFP open reading frame; BGH polyA—Bovine Growth Hormone polyadenylation signal. The names of the dual fluorescence constructs indicate only the presence of DIVACs, the RFP gene and sequence repeats. (B) FACS dot plots of representative subclones derived from primary transfectants. The number of the plot and the name of the transfected construct are indicated above. Plots of subclones in which the AID expression cassette have been deleted are labeled AID−. Gates and the percentage of gated cells are indicated within the plots. (C) Graphs for each gate showing the percentages of gated cells for all subclones of each independent transfectant. The median percentage of gated cells is indicated by the bar and numerically displayed above the graph for each transfectant.
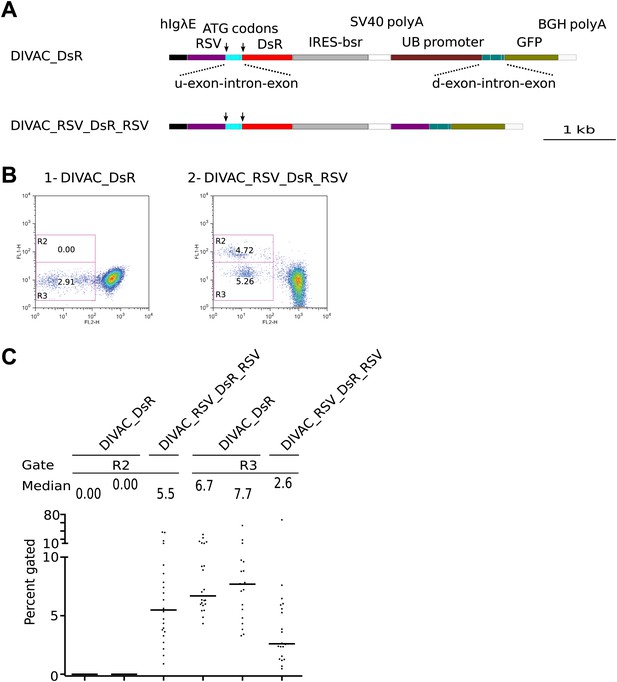
FACS analysis of DT40 transfectants expressing dual DsR/GFP fluorescence reporter constructs.
(A) Maps of the constructs. (B) FACS dot plots of representative subclones derived from primary transfectants. (C) Graphs for each gate showing the percentages of gated cells for all subclones of each independent transfectant. The median percentage of gated cells is indicated by the bar and numerically displayed above the graph for each transfectant.
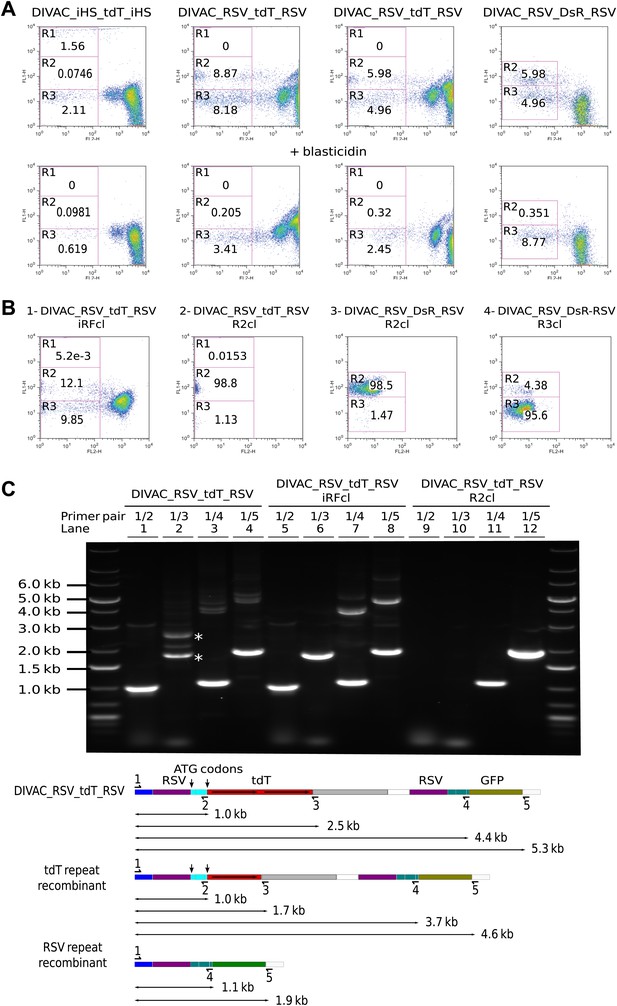
Homologous recombination of sequence repeats in gated cells.
(A) FACS profiles of primary transfectants 20 days after transfection. Cells analyzed in the upper row were cultured in the absence of blasticidin, those in the lower row in the presence of blasticidin. (B) Representative FACS profiles of subclones derived from a cell of intermediate red fluorescence or from R2- and R3-gated cells. The transfected constructs are indicated above the profiles by name, with the origin of the precursor cell indicated by the suffix. (C) Top: agarose gel electrophoresis of PCR products amplified from DNA of the DIVAC_RSV_tdT_RSV transfectant and two of its subclones. The first subclone is derived from a cell of intermediate red fluorescence, the second from a R2 cell. The primers used are indicated above the numbered lanes. Bands representing amplifications of the un-rearranged and rearranged tdT genes are marked by asterisks in lane 2. Below: the diagram shows the positions of the primers and the expected sizes of the PCR products for the transfected construct and its recombinants. The increased GFP expression of recombinants is indicated by color changes of the rectangle representing the GFP open reading frame.
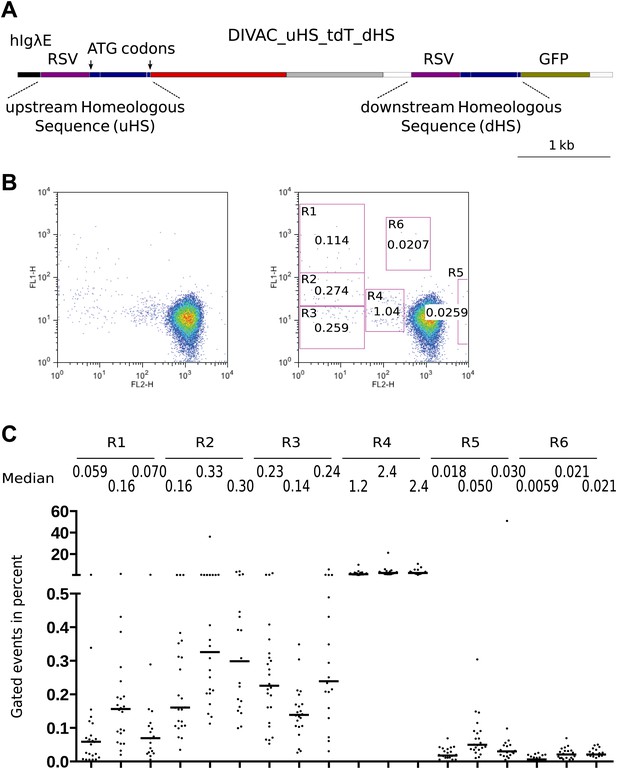
FACS profiles of transfectants of the DIVAC_uHS_tdT_dHS construct.
(A) Diagram of the construct. The upstream and downstream homeologous sequence repeats (uHS and dHS, respectively) are highlighted. (B) FACS profile of a representative subclone. (C) Graphs showing the percentages of gated cells for subclones of three independent transfectants. The median percentage of gated cells of all subclones is indicated by the bar and numerically displayed above the graph for each transfectant.
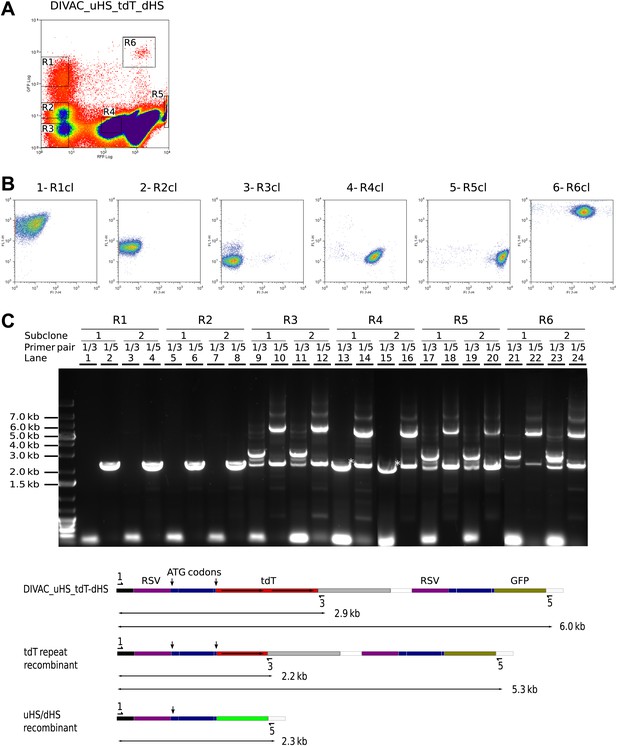
Recombination of the uHS/dHS repeat in R1 and R2 cells of the DIVAC_uHS_tdT_dHS transfectant.
(A) FACS profile of a DIVAC_uHS_tdT_dHS transfectant showing the gates used for preparative sorts. (B) FACS plots of representative subclones. The gate from which the precursor cell of the subclone is derived is shown above the plot. (C) Agarose gel electrophoresis of PCR products amplified from DNA of subclones which were derived for gated cells as indicated on top of the gel image. The primer pairs used for the amplifications are shown above the lanes. The lower scheme shows the positions of the primers and the expected sizes of the PCR products for the transfected construct and its recombinants.
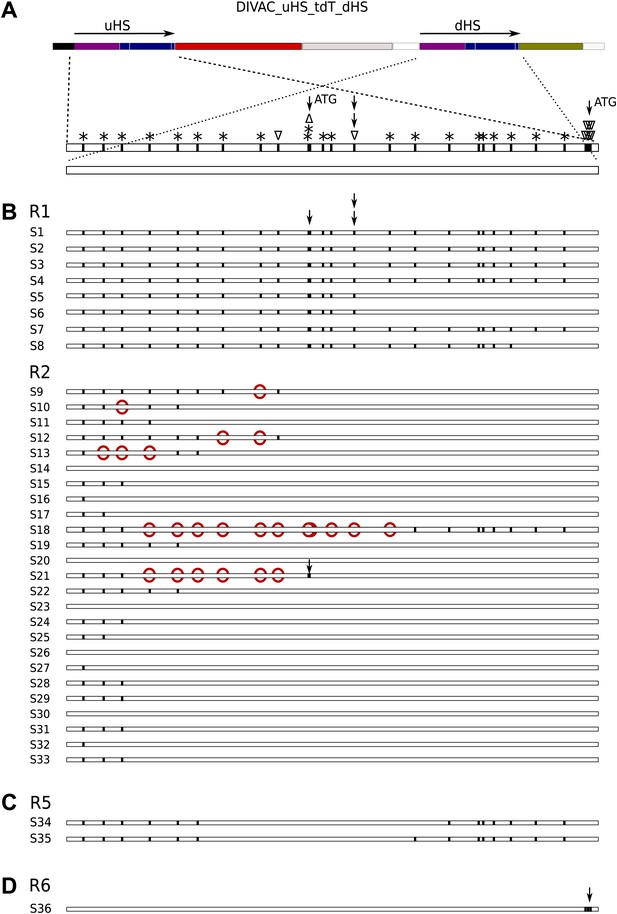
Sequences derived from gated cells of the DIVAC_uHS_tdT_dHS transfectant.
(A) Map of the construct showing the polymorphisms of the aligned uHS and dHS sequences. The types of polymorphism are coded above the uHS sequence. Asterisks indicate nucleotide substitutions while triangles pointing up and down indicate single nucleotide insertions and deletions, respectively, within uHS. The positions of the first and second ATG start codon of uHS are highlighted by single arrows, and the position of the single nucleotide deletion that puts the first ATG in frame with tdT is indicated by stacked arrows. In all parts of the figure vertical lines within the sequence bars indicate uHS specific sequence. (B) Schematic representation of the sequences of uHS/dHS recombinants amplified from sorted R1 and R2 cells. uHS specific nucleotides downstream of dHS nucleotides are marked by red circles. (C) uHS sequences amplified from sorted R5 cells. (D) A dHS sequence amplified from a R6 subclone.
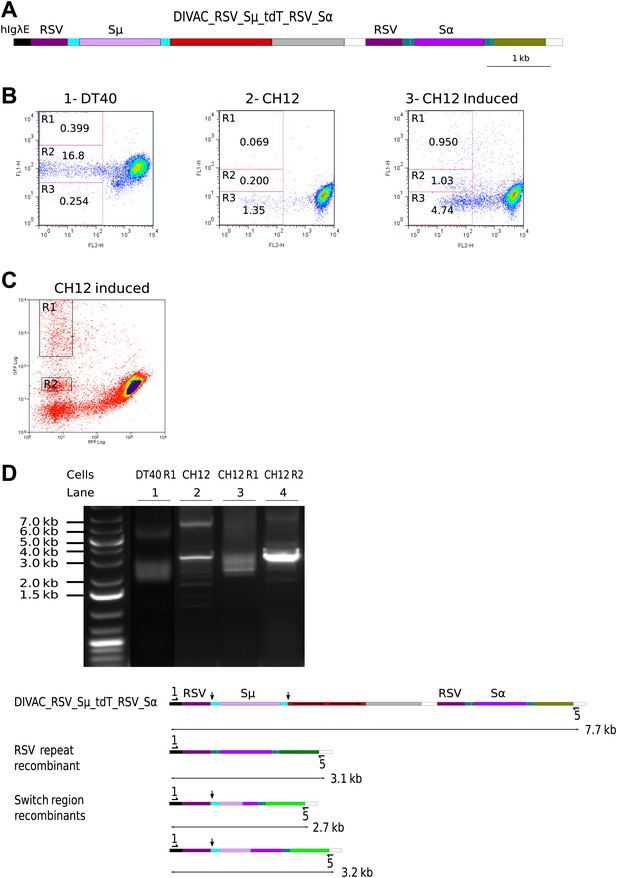
RSV repeat and class switch recombination after transfection of CH12 cells.
(A) Diagram of the CH12 DIVAC_RSV_Sμ_tdT_RSV_Sα construct. Sμ and Sα—portions of the murine Sμ and Sα switch regions. (B) FACS profile of a representative DT40 transfectant as well as an uninduced and induced CH12 transfectant. (C) FACS profile of the induced CH12 transfectant showing the gates of the preparative sorts. (D) Top: agarose gel electrophoresis of PCR products amplified by the 1/5 primer pair from DNA of sorted DT40 R1 cells, the induced CH12 transfectant and sorted CH12 R1 and CH12 R2 cells. Bottom: the diagrams show the positions of the primers and the expected sizes of the PCR products for the transfected construct and its recombinants.
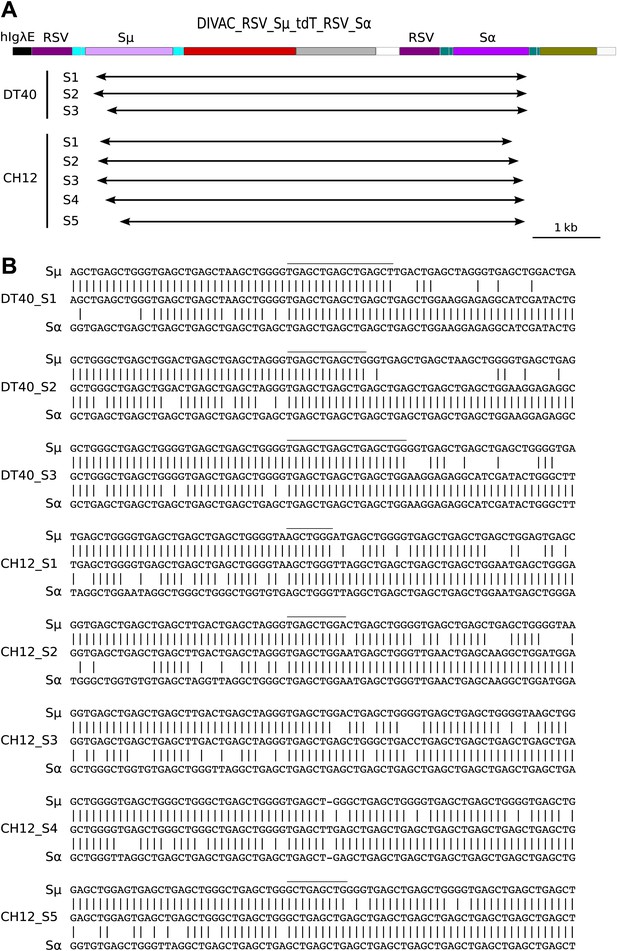
Sequences of switch region recombinants recovered from sorted DT40 R1 and CH12 R1 cells.
(A) Deletions induced by the joining of Sμ and Sα switch regions are indicated below in the map of the DIVAC_RSV_Sμ_tdT_RSV_Sα construct by horizontal arrows. (B) Switch region junctions are aligned to the sequence of Sμ and Sα. Aligned Sμ and Sα sequences of the construct are shown above and below, respectively, the sequence of each switch recombinant. Junctional microhomologies are marked by a line above the sequence.
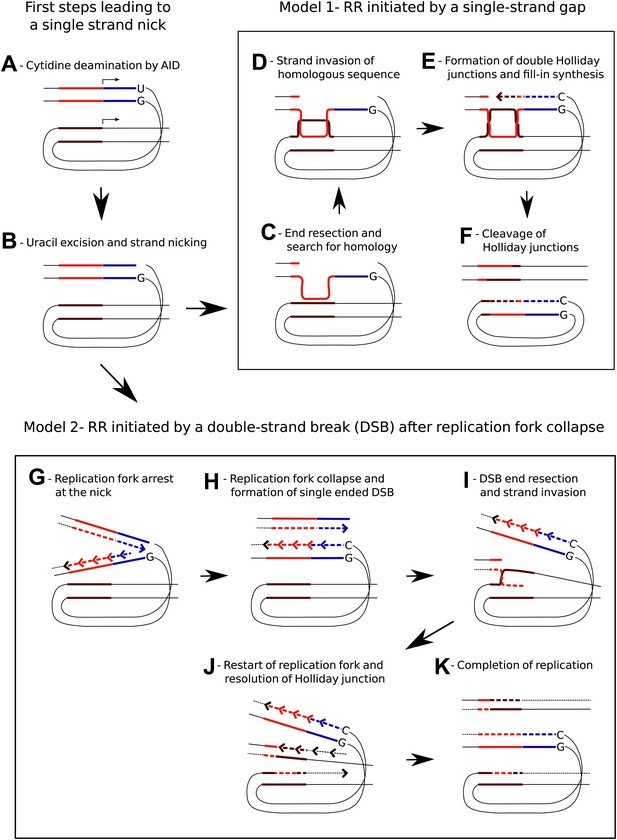
Models for AID induced Recombination of Repeats (RR) assuming initiation of homologous recombination by a single strand gap or a double strand break.
(A) The diagrams show two neighboring genes containing direct sequence repeats marked in red and brown. The transcription start sites and the direction of transcription are indicated by horizontal arrows. Deamination of a cytidine by AID within the transcribed sequence of the first gene, marked in blue, leads to an uracil/guanidine base pair (‘U’ opposite to ‘G’). (B) Removal of the uracil and cleavage of the abasic site results in a single-strand nick that is postulated to be a common intermediate for the two models. (C) The first model (Model 1, C–F) assumes that 5′ to 3′ resection of the nick produces a gapped DNA duplex and that the unpaired, continuous strand initiates the search for homology. (D) D-loop formation at the downstream repeat sequence. (E) Following strand exchange DNA synthesis shown by the dashed lines fills in the gap in the DNA duplex. (F) Cleavage of the Holliday junctions in one plane creates a chromosome containing a deletion and a circular DNA molecule of the deleted sequence. Cleavage of the Holliday junctions in the other plane would result in a chromosome with no deletion (not shown). (G) The second model (Model 2, G–K) assumes replication arrest at the AID-induced nick. (H) The nick is converted into a single ended DSB due to replication fork collapse. (I) Upon resection the DSB erroneously re-initiates the replication fork at the position of the downstream repeat sequence. (J) The Holliday junction is cleaved and replication continues. (K) Different sister chromatids, one carrying a deletion and one without a deletion, are produced by completion of replication.





