How lamina-associated polypeptide 1 (LAP1) activates Torsin
Figures
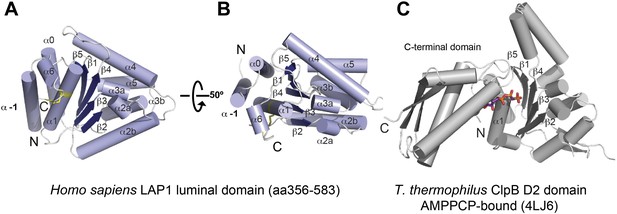
Crystal structure of human LAP1.
(A) Crystal structure of the luminal domain of LAP1 from Homo sapiens. The helices are numbered according to the nomenclature used for AAA+ proteins (Erzberger and Berger, 2006). A disulfide bridge (yellow) attaches the C terminus to helix α1. (B) Same as (A), but rotated around the x-axis by 50°. (C) Crystal structure of the AMPPCP-bound ClpB-D2 domain from Thermus thermophilus (4LJ6; Zeymer et al., 2014) in the same orientation as LAP1 in (A) revealing the striking topological similarity. In comparison to ClpB, LAP1 does not bind a nucleotide and has no C-terminal domain.
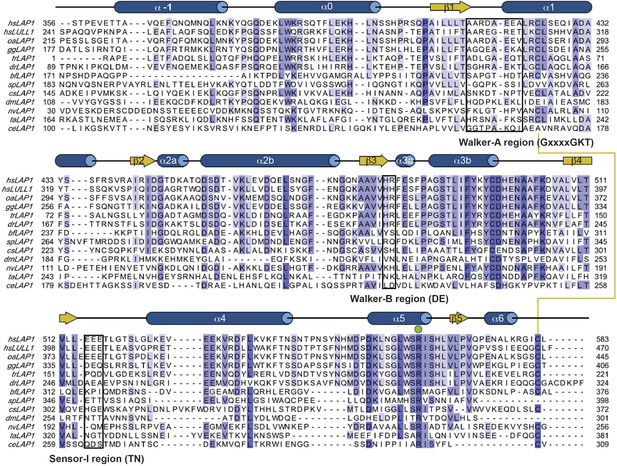
Phylogenetic analysis of LAP1 homologs.
Sequence alignment of phylogenetically diverse sequences of the luminal domain of LAP1 and LULL1. Residues are color-coded from dark blue (most conserved) to white (not conserved). Secondary structure elements of human LAP1 are shown above the sequences. The conserved disulfide bond is indicated with a yellow line. The strictly conserved arginine (‘R-finger’) is marked by a green circle.
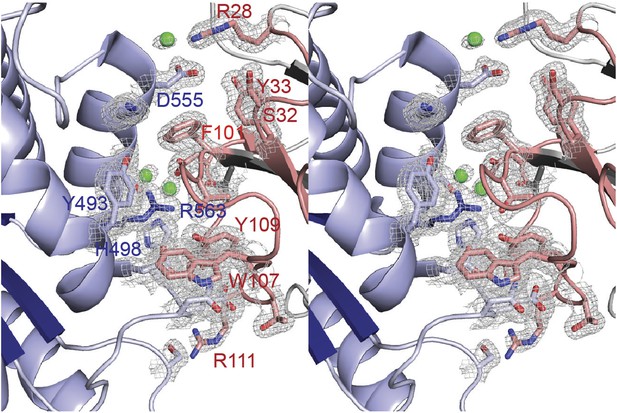
Representative 2Fo-Fc electron density of the final model.
Stereo view of the binding site of VHH-BS1 (gray) with LAP1 (blue). Contacting residues between LAP1 and the variable loops of VHH-BS1 (red) are shown as sticks and are labeled. Waters, with direct contact to both binding partners, are in green. The 2Fo-Fc map is contoured at 1σ.
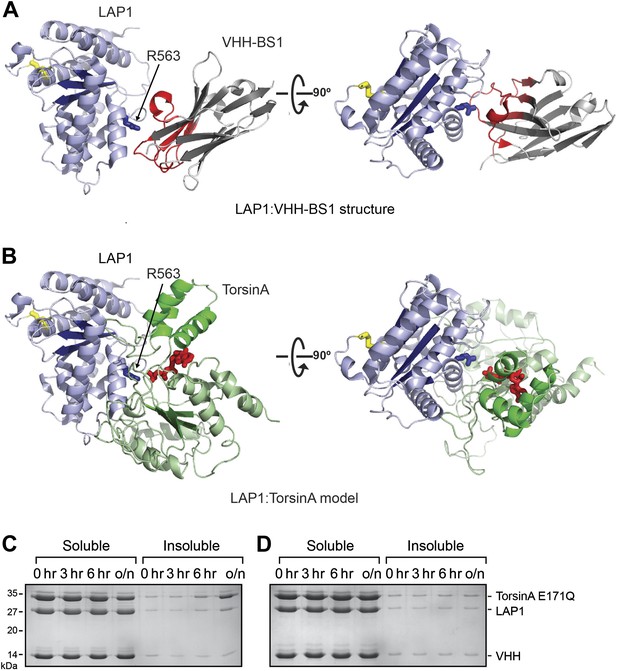
VHH-BS1 competes with TorsinA for LAP1 binding.
(A) Overall structure of the LAP1-VHH-BS1 complex. LAP1 is in light blue, VHH-BS1 in gray. The variable loops, responsible for high affinity interaction with LAP1, are colored in red. The binding site on LAP1 is centered around arginine 563 (blue sticks). (B) Model of LAP1 (light blue) interacting with TorsinA (green) in a putative heterohexameric ring assembly in the same orientations as in (A). TorsinA-bound ATP molecule is in red. Note the overlap between the VHH-BS1 and the putative TorsinA binding site on LAP1. (C and D) In vitro precipitation assay shows that VHH-BS1 competes with TorsinA(E171Q) for LAP1 binding. Preformed TorsinA(E171Q)-LAP1 complex was incubated for several hours with VHH-BS1 (C) or a control VHH (D). The mixtures were centrifuged at the indicated times to separate soluble from insoluble protein. The samples were analyzed by SDS-PAGE. Due to the competition with VHH-BS1, TorsinA(E171Q) is released from LAP1. Since free TorsinA(E171Q) is insoluble, the process of the reaction can be monitored.
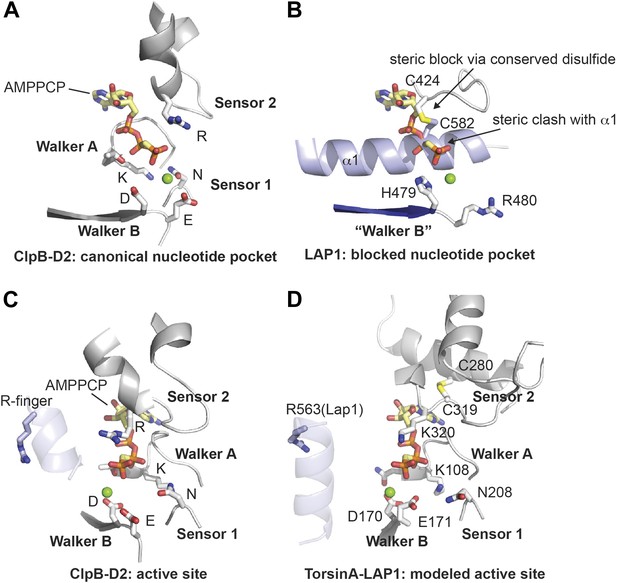
Nucleotide binding site.
(A and B) Comparison of the nucleotide binding pocket in ClpB-D2 (A) with the equivalent region in LAP1 (B). The nucleotide sensing elements of canonical ATPases are indicated. The nucleotide in (B) is modeled to illustrate how LAP1 blocks binding of it. (C and D) To activate ATP hydrolysis, an arginine residue (R-finger) from the neighboring protomer in the typical hexameric ring assembly is necessary. For ClpB-D2, this is R747 (light blue) (C). Modeled as a heterohexameric LAP1-TorsinA assembly, the strictly conserved R563 in LAP1 is positioned as an R-finger to point into the nucleotide-binding pocket of a neighboring TorsinA protomer (D). Residues important for nucleotide interaction are labeled in the TorsinA model.
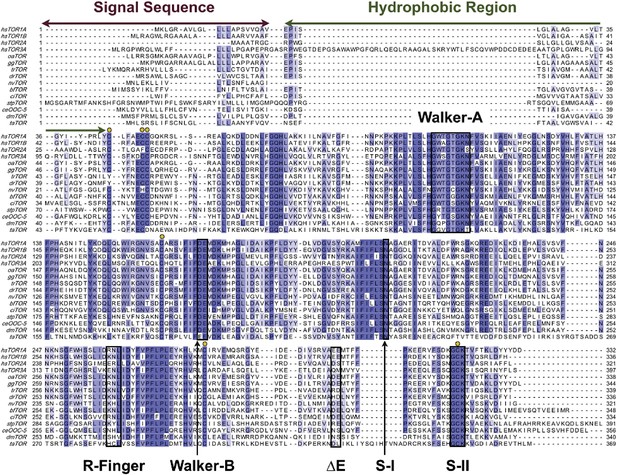
Phylogenetic analysis of Torsin.
Sequence alignment of phylogenetically diverse sequences of Torsin. Residues are color-coded from dark blue (most conserved) to white (not conserved). The canonical nucleotide interaction motifs, and other Torsin-specific regions, are labeled. Conserved cysteines are marked with yellow circles. Note that within the R-finger region, Torsin has no conserved arginine.
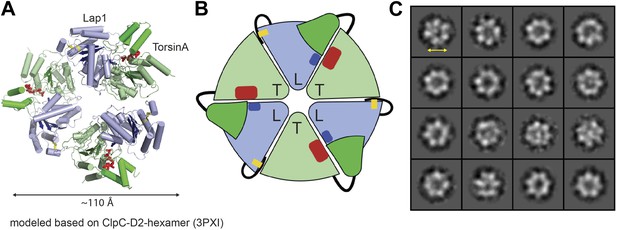
Heterohexameric ring assembly.
(A) Heterohexameric model of alternating LAP1 (light blue) and TorsinA (green hues), based on the hexameric ring of ClpC-D2 domains (3PXI; Wang et al., 2011). The C-terminal domain of Torsin is colored bright green. An ATP molecule (red) is modeled into the TorsinA nucleotide binding pocket. The conserved arginine finger in LAP1 is in blue. The disulfide bridge within LAP1 is in yellow. (B) Schematic diagram of (A), same coloring scheme. (C) A montage of representative particle classes of the TorsinA(E171Q):LULL1 complex obtained from negatively stained particles are shown. The particles are consistent with a toroidal hexameric conformation as shown in (A and B). The particle diameter is approximately 120 Å (double arrow).
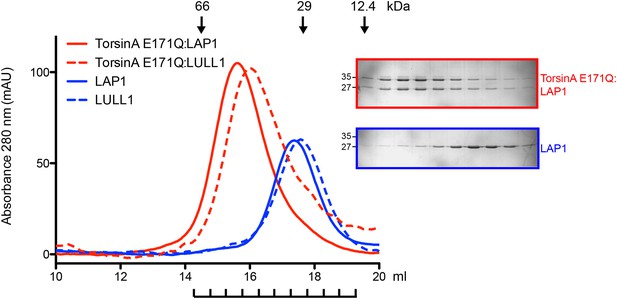
Analytical gel filtration of TorsinA(E171Q):LAP1 and TorsinA(E171Q):LULL1 complexes.
Elution profiles of TorsinA(E171Q):LAP1 (red) and TorsinA(E171Q):LULL1 (red dashes) are compared with uncomplexed LAP1 (blue) and LULL1 (blue dashes), respectively. The experiment was performed on a Superdex S200 HR10/300 column. Elution volumes of standard globular protein markers are displayed. SDS-PAGE analysis of peak fractions is shown. The fractions are indicated below the chromatogram. The TorsinA:LAP1 and TorsinA:LULL1 protein complexes elute at positions consistent with a heterodimer. SDS-PAGE analysis indicates a 1:1 stoichiometry for the complexes.
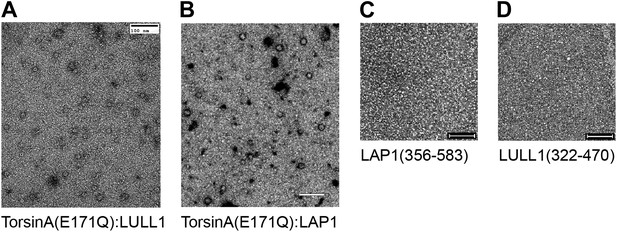
Negative-stain micrographs.
Protein samples were negatively stained with uranyl acetate and analyzed by electron microscopy. All samples were measured at closely comparable concentrations. Scale bars represent 100 nm. While ring-like structures are visible in the TorsinA(E171Q):LULL1 and TorsinA(E171Q):LAP1 samples (A and B), no discernible objects are seen on the micrographs of uncomplexed LAP1 and LULL1 (C and D). This indicates that LAP1 and LULL1 do not form homohexameric rings in the absence of TorsinA.
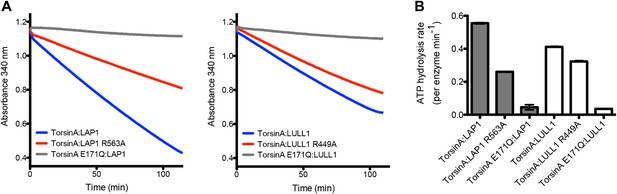
ATPase assay.
(A) TorsinA:LAP1 and TorsinA:LULL1 complexes were tested for ATPase activity in a coupled ADP/NADH assay, where the oxidation of NADH is monitored spectrometrically (Norby, 1988). Shown are the oxidation rate plots, from which the ATP hydrolysis rates are calculated using linear regression. Reactions were performed in triplicate. (B) Bar graph representation of ATPase assay results shown in (A).
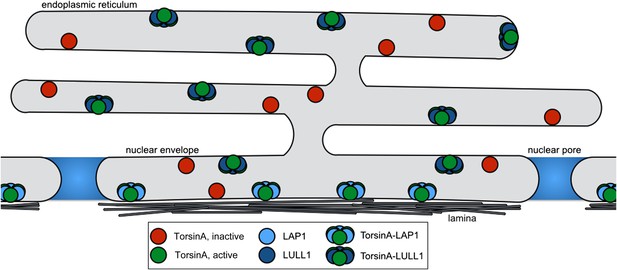
Model for Torsin activation and localization.
LAP1 (light blue) is localized to the nuclear envelope due to its interaction with the nuclear lamina, while LULL1 (dark blue) is found throughout the endoplasmic reticulum. Both proteins can bind inactive TorsinA (red) and target it to their respective locations. Both LAP1 and LULL1 activate TorsinA (green) when assembled into heterohexameric rings.
Tables
X-ray data collection and refinement statistics
| Protein | Human LAP1356–583-VHH-BS1 complex |
| PDB ID | 4TVS |
| Data collection | |
| Space group | P21 |
| a, b, c (Å) | 69.79, 74.02, 85.43 |
| α, ß, γ (°) | 90, 108.8, 90 |
| Wavelength (Å) | 1.2548 |
| Resolution range (Å)* | 66.1–1.60 (1.66–1.60) |
| Total reflections | 630,312 (46,516) |
| Unique reflections | 105,976 (10,195) |
| Completeness (%) | 97.6 (94.3) |
| Redundancy | 5.9 (4.6) |
| Rsym (%) | 9.1 (141.2) |
| Rp.i.m. (%) | 4.0 (68.6) |
| I/σ | 12.5 (1.1) |
| CC1/2 (%) | 99.8 (54.7) |
| Refinement | |
| Resolution range (Å) | 66.1–1.60 |
| Rwork (%) | 18.1 |
| Rfree (%) | 22.9 |
| Coordinate error (Å)† | 0.24 |
| Number of reflections | |
| Total | 105,565 |
| Rfree reflections | 2810 |
| Number of non-hydrogen atoms | |
| Protein | 5520 |
| Ligands | 29 |
| Water | 612 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 1.66 |
| B-factors (Å2) | |
| Protein | 39.3 |
| Ligands | 56.4 |
| Water | 47.8 |
| Ramachandran (%)‡ | |
| Favored (%) | 98.3 |
| Outlier (%) | 0.0 |
| Clashscore | 5.65 |
| MolProbity score‡ | 1.29 |
| MolProbity percentile‡ | 96th |
-
*
Numbers in brackets refer to the highest resolution shell (10% of all reflections).
-
†
Maximum likelihood based (as determined by PHENIX; Adams et al., 2010).
-
‡
As determined by MolProbity (Chen et al., 2010).





