The Drosophila F-box protein Fbxl7 binds to the protocadherin Fat and regulates Dachs localization and Hippo signaling
Figures
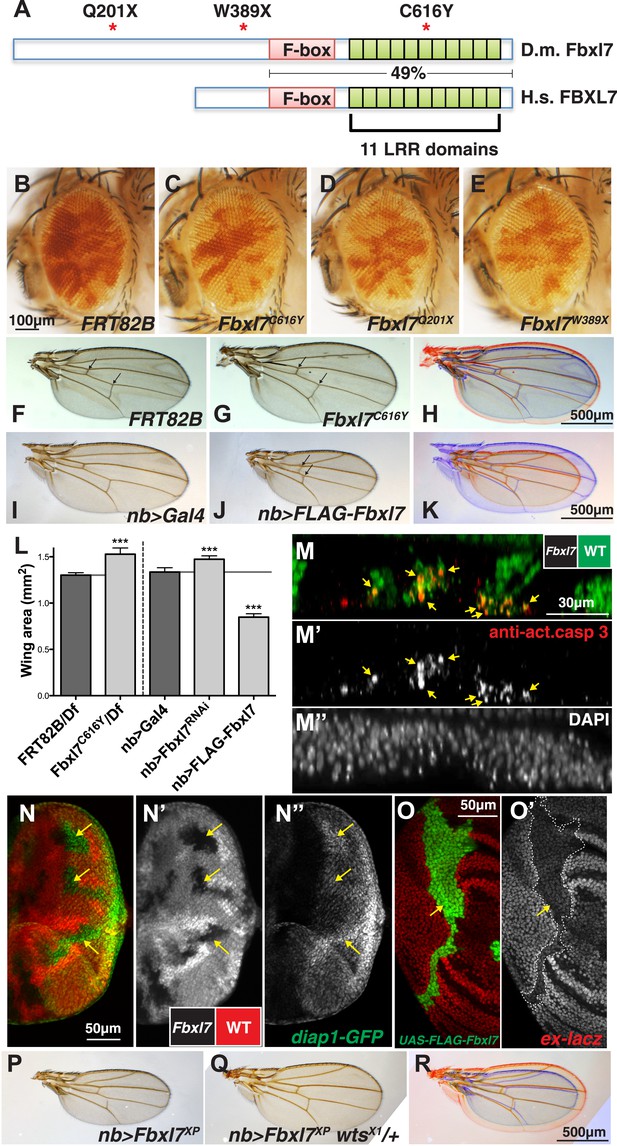
Fbxl7 negatively regulates growth through the Hippo pathway.
(A) Protein model of Drosophila Fbxl7 and Human FBXL7 showing the three alleles identified (red asterisks), F-box, and 11 Leucine Rich Repeat (LRR) domains. The two proteins have 49% amino acid identity throughout the F-box and LRR domains. (B–E) Mosaic adult eye assay. Heterozygous and wild-type cells have red pigment and homozygous mutant cells lack pigment. (B) Control mosaic eye. (C) Fbxl7C616Y, (D), Fbxl7Q201X and (E) Fbxl7W389X mosaic eyes are composed of more mutant cells. (F–K) Adult wings with overlays. Arrows indicate anterior and posterior crossveins. Compared to (F) FRT82B control wings, (G) Fbxl7C616Y homozygous wings are larger and crossveins are closer. (H) Merge shows F in blue and G in red. Compared to (I) nubbin-Gal4 (nb-Gal4) control wings, (J) nb>FLAG-Fbxl7 overexpressing wings are smaller and crossveins are closer. (K) Merge shows I in blue and J in red. (L) Quantification of wing area from Fbxl7 loss-of-function, RNAi (JF01515), and overexpression. n ≥ 20 wings, ***p ≤ 0.001, error bars show SD. (M–M″) Cell competition assay in the mosaic eye imaginal disc. (M) Wild-type cells are marked by GFP (green), while Fbxl7 mutant cells are GFP negative. (M′) Activated caspase-3 (red) is detected in dying cells that are GFP positive (arrows). (M″) DAPI shows all nuclei. (N–N″) Mosaic eye imaginal disc with diap1-GFP (green) reporter. (N–N′) Wild-type cells are marked with RFP (red) and Fbxl7 mutant cells are RFP negative. (N″) Mutant clones show higher levels of diap1-GFP (arrows). (O–O′) Mosaic wing imaginal disc with ex-lacZ reporter (red). A clone overexpressing FLAG-Fbxl7 (green, cells marked by EGFP) has lower levels of ex-lacZ (arrow). (P–R) Wing size genetic interaction assay. Compared to (P) nb>Fbxl7XP alone, (Q) reducing the dosage of wts partially rescues the small wing phenotype. (R) Merge shows P blue and Q in red.
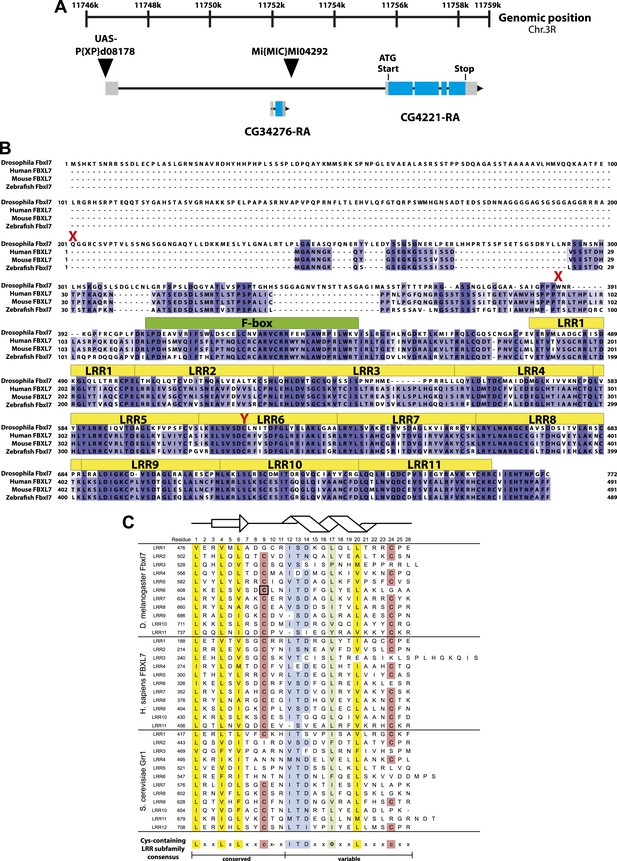
Fbxl7 gene and protein models.
(A) Fbxl7 gene model. CG4221-RA is the only predicted isoform. Thin black lines are introns, gray boxes indicate non-coding exons, and blue boxes indicate coding exons. CG34276 is located in the first intron of Fbxl7. P{XP}d08178 is inserted immediately upstream of the Fbxl7 transcript and contains UAS sequences to drive ectopic expression of Fbxl7. Mi{MIC}MI04292 is inserted in the first intron of Fbxl7. (B) Fbxl7 protein alignment of Drosophila Fbxl7 (NP_650512.1), Human FBXL7 (NP_036436.1), Mouse FBXL7 (NP_795933.2), and Zebrafish Fbxl7 (NP_001073511.1). Alignment performed with Clustal Omega and exported with Jalview software. Conserved F-box domains are shown in green and 11 Leucine Rich Repeat (LRR) domains are shown in yellow. Drosophila Fbxl7 mutations indicated in red. (C) Alignment of Cys-containing LRR subfamily domains from Drosophila Fbxl7, Human FBXL7, and S. cerevisiae Grr1 (NP_012623.1). LRRs identified using SMART software (Letunic et al., 2012). Black box indicates the Drosophila Fbxl7C616Y amino acid mutation that affects a conserved cysteine. Structural model shows the predicted LRR β-sheet (arrow) and α-helix (helix) modeled after (Hsiung et al., 2001). Consensus sequence definitions according to Bella et al. (2008) and Kobe and Deisenhofer (1994).
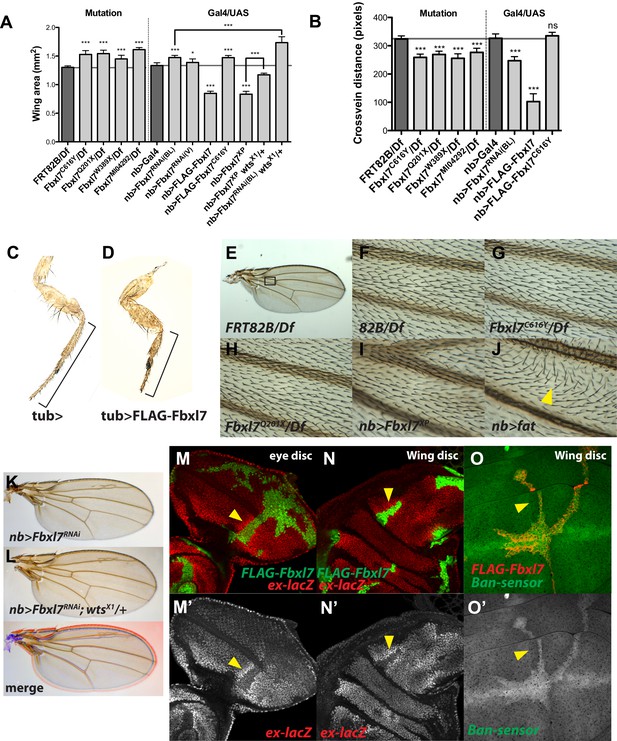
Additional Fbxl7 mutant phenotypes.
(A–B) Quantification of adult wing area and cross vein distance for different Fbxl7 loss of function and overexpression phenotypes. n ≥ 10 wings. Significance calculated with 1way ANOVA followed by Tukey's test. ***p ≤ 0.001, *p ≤ 0.05. Error bars indicate SD. (C–D) Adult male prothoracic legs from control or overexpressing FLAG-Fbxl7. Black bracket shows shortening of tibia and tarsus segments. (E–J) Wing hair polarity in the region between veins L3 and L4 and proximal to the anterior cross vein (black box in E). (J) Overexpressing fat causes wing hairs to change direction (yellow arrowhead). (K–L) Increased adult wing size from RNAi knockdown of Fbxl7 in the wing is enhanced by wts heterozygosity. Merge shows K in blue and L in red. (M–O) Confocal slices of imaginal discs with clones overexpressing FLAG-Fbxl7 and assessing Hippo pathway reporters ex-lacZ and Tub-EGFP.ban (‘bantam sensor’). ex-lacZ positively reports Yki activity, whereas bantam-sensor inversely reports bantam activity (bantam, a microRNA, is a transcriptional target of Yki). (M–N) Cells overexpressing FLAG-Fbxl7 (marked by EGFP, green) have lower levels of ex-lacZ (red), and neighboring wild-type cells have higher levels of ex-lacZ (arrowhead). (O) Cells overexpressing FLAG-Fbxl7 (marked by myr-RFP, red) have higher levels of bantam-sensor (green), and neighboring wild-type cells have lower levels of bantam-sensor (yellow arrowhead).
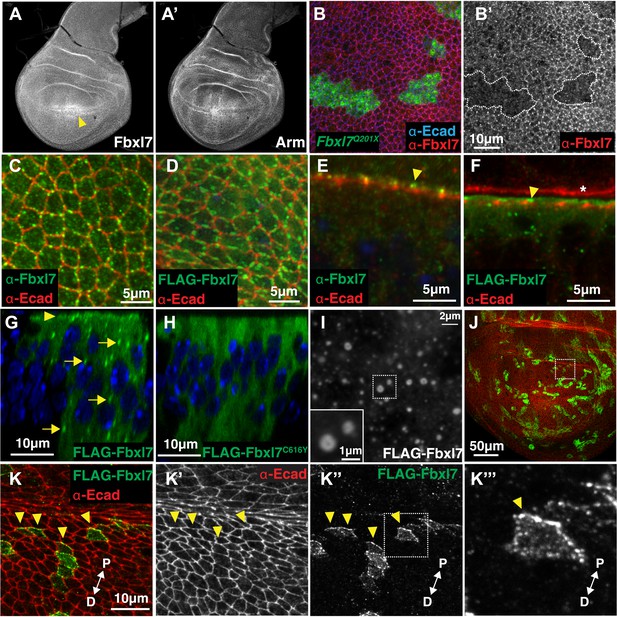
Fbxl7 is localized to apical membrane, cytoplasmic puncta, and the proximal side of planar polarized cells.
Confocal slice of (A) endogenous Fbxl7 and (A′) Armadillo (Arm) in the wing imaginal disc. Arrow indicates enrichment of Fbxl7 at the dorso-ventral boundary. (B–B′) A confocal slice through the apical surface of wing disc cells. Fbxl7 (red) accumulates at the apical membrane and is lost from MARCM Fbxl7Q201X clones (green). (C–F) Endogenous Fbxl7 and expressed FLAG-Fbxl7 (green) are localized to apical puncta aligned with cell edges marked by E-cadherin (E-cad) (red). (C–D) Confocal slices through the apical surface of wing disc cells. (E–F) Confocal slice through folds in the wing disc. Fbxl7 is apical to E-cad (arrowheads). (F) Asterisk indicates adjacent fold that does not express FLAG-Fbxl7. (G–H) Confocal Z-slice through the wing disc with clones of cells expressing FLAG-Fbxl7 or FLAG-Fbxl7C616Y (green). Nuclei are shown with DAPI (blue). (G) FLAG-Fbxl7 localizes to apical membrane (arrowhead) and cytoplasmic puncta (arrows), whereas (H) FLAG-Fbxl7C616Y shows diffuse cytoplasmic localization. (I) Confocal section through peripodial membrane showing FLAG-Fbxl7 localization to hollow puncta. Inset shows higher magnification of outlined box. (J–K‴) Confocal slice of the wing disc pouch stained for E-cad (red) with clones expressing FLAG-Fbxl7 (green). (K–K″) Magnified region from box in J, showing FLAG-Fbxl7 enriched on proximal membrane (arrowheads). (K‴′’) Magnified region from box in K‴. D = distal, P = proximal.
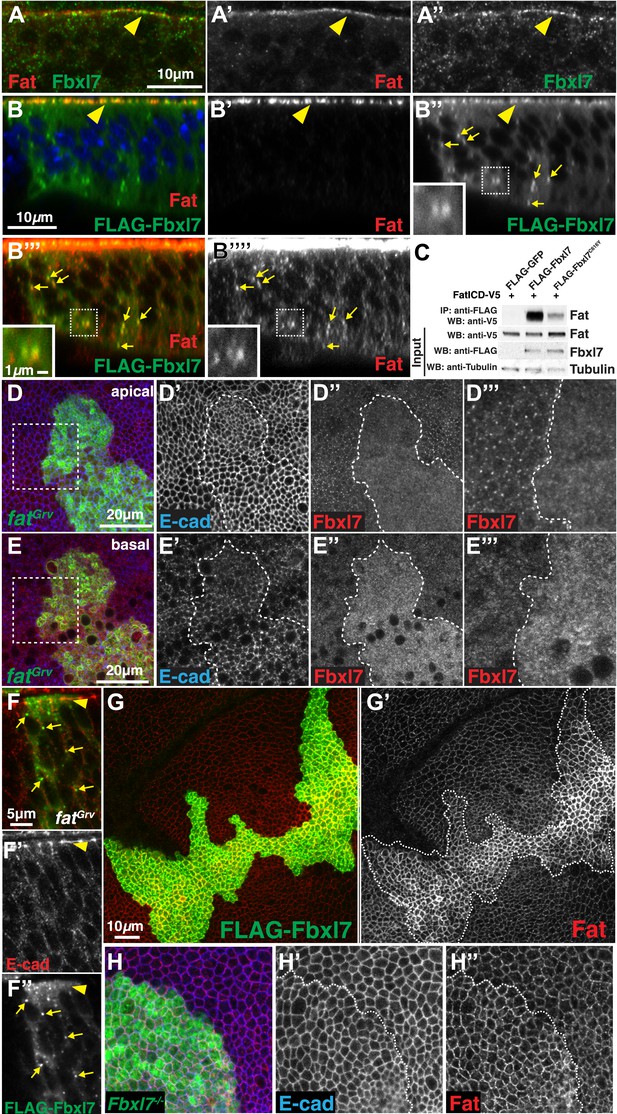
Fbxl7 physically interacts with Fat and regulates its apical localization.
(A–A″) Confocal slice through a wing disc fold showing endogenous Fbxl7 (green) and Fat (red) co-localize at apical membrane (arrowhead). (B–B‴′) Confocal Z-section showing FLAG-Fbxl7 (green) and Fat (red) co-localize at (B–B″) apical membrane (arrowhead) and (B″–B‴′) cytoplasmic puncta (arrows). (B″–B‴′) Inset shows magnification of puncta. B‴′ uses higher gain settings than B′ to visualize Fat in puncta. (C) Co-immunoprecipitation experiment in S2 cells. FatICD-V5 pulls down with FLAG-Fbxl7, whereas pulldown is reduced with FLAG-Fbxl7C616Y. (D–D‴) Confocal slice of the wing disc at the apical surface. Apical Fbxl7 (red) localization is lost from MARCM fatGrv clones (green), whereas (D′) E-cad (blue) localization is unchanged. (D‴) shows magnification of the box in D. (E–E‴) A basal confocal slice through the same clone in D, showing increased cytoplasmic levels of Fbxl7. (F–F″) Confocal slice through a fold showing a MARCM fatGrv clone (GFP marker not shown) which expresses FLAG-Fbxl7 (anti-Flag, green). (F′) E-cad (red) marks apical membrane. FLAG-Fbxl7 is not apically localized in fatGrv clones (arrowhead), but does localize to cytoplasmic puncta (arrows). (G–G′) Confocal slice through the apical surface of a disc overexpressing FLAG-Fbxl7 (green) in clones. (G′) Apical Fat (red) levels are elevated within the clone. (H–H″) Confocal slice through the apical surface with a MARCM Fbxl7Q201X clone (green) showing (H′) no change in levels of apical E-cad (blue) and (H″) slightly elevated levels of apical Fat (red).
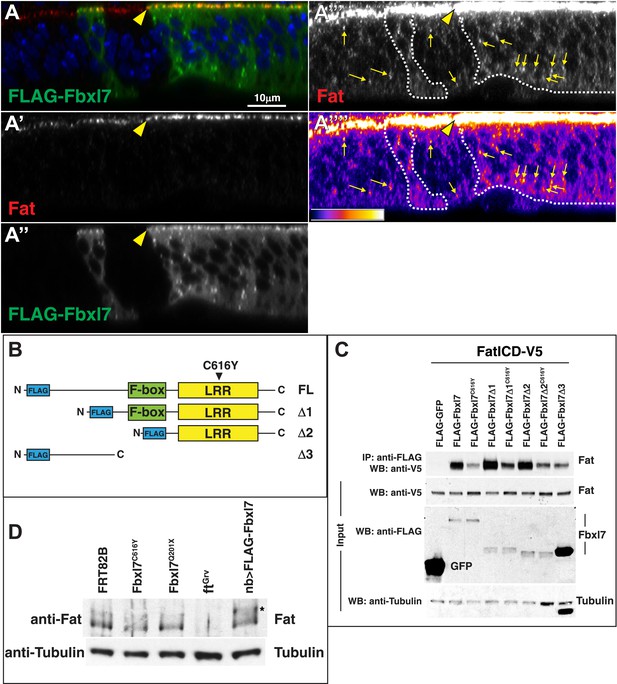
Additional analysis of the relationship between Fbxl7 and Fat.
(A) A confocal z-section through a wing disc with clones overexpressing FLAG-Fbxl7 and staining for FLAG (green) and Fat (red). Apical membrane is towards the top of the image. Fat and FLAG-Fbxl7 colocalize at apical membrane (yellow arrowhead). (A‴–A‴′) Higher gain settings to observe Fat cytoplasmic punctae and heat map of fluorescence intensity. Fat localizes to punctae in both wild-type cells and FLAG-Fbxl7 expressing cells (yellow arrows). Cytoplasmic Fat is slightly elevated in FLAG-Fbxl7 expressing cells. (B) Schematics of FLAG-Fbxl7 truncation constructs. (C) Western blots showing results of co-immunoprecipitation experiments from S2 cells expressing indicated transfected plasmids. FatICD co-immunoprecipitates with full length FLAG-Fbxl7 as well as the LRR domain. The Fbxl7C616Y protein reduces association with FatICD. The N-terminal domain of Fbxl7 can weakly associate with FatICD. FLAG-Fbxl7Δ3 protein is found at higher levels than other Fbxl7 proteins despite transfecting the same amount of plasmid and loading the same amount of total protein. (D) Western blots showing endogenous Fat protein from wing disc lysates of indicated genotypes. The asterisk indicates higher molecular weight Fat in discs overexpressing FLAG-Fbxl7.
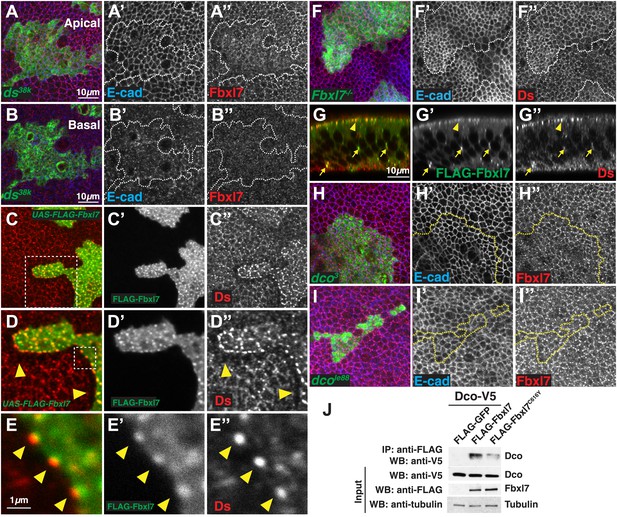
Relationship between Fbxl7 and the Fat pathway proteins Ds and Dco.
(A–A″) Confocal slice through the apical surface of a disc with MARCM ds38k clones (green) showing disturbed localization of Fbxl7 (red). E-cad staining is not altered (blue) (B–B″) A basal confocal slice through the same clone in A, showing no change in Fbxl7 cytoplasmic levels. (C–E″) Confocal slice through the apical surface of a disc with FLAG-Fbxl7 overexpressing clones (green) and stained for Ds (red). (C–D″) Apical Ds levels appear higher and more punctate in FLAG-Fbxl7 expressing clones. Wild-type cells immediately adjacent to the clone have reduced apical Ds (arrowheads). (E–E″) Ds and FLAG-Fbxl7 puncta are aligned on either side of the clone boundary (arrowheads). (F–F″) Apical confocal slice of a disc containing MARCM Fbxl7Q201X clones (green) and stained for Ds (red) and E-cad (blue). Ds levels are normal or slightly elevated, in clones. (G–G″) Confocal Z-section of a clone expressing FLAG-Fbxl7 (green) and stained for Ds (red). Both are localized to apical membrane (arrowhead) and frequently co-localize in cytoplasmic puncta (arrows). (H–I″) Apical confocal slice of MARCM dco3 or dcole88 clones (green) and staining for Fbxl7 (red) and E-cad (blue). Apical Fbxl7 levels are unchanged in (H–H″) dco3 and (I–I″) dcole88 clones. (J) Co-immunoprecipitation experiment in S2 cells. Dco-V5 pulls down with FLAG-Fbxl7, whereas pulldown is reduced with FLAG-Fbxl7C616Y.
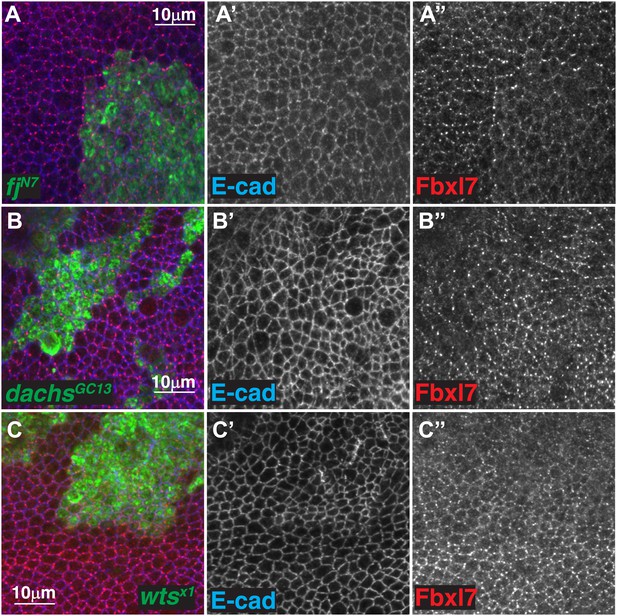
Additional analysis of the relationship between Fbxl7 and Fat pathway proteins.
(A) Confocal slice through the apical membrane of wing disc cells bearing MARCM fjN7 clones (green) and antibody stained for Fbxl7 (red) and E-cad (blue). Fbxl7 apical localization is partially disrupted. (B–C″) Confocal slices through the apical membrane of wing disc cells bearing MARCM clones (green) and antibody stained for Fbxl7 (red) and E-cad (blue). Fbxl7 apical localization is normal in (B–B″) dachsGC13 or (C–C″) wtsX1 clones.
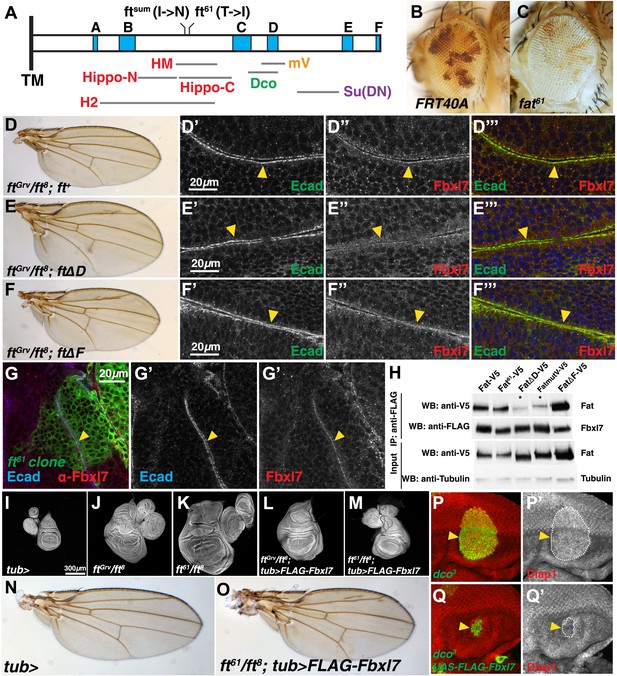
Fbxl7 functions in one of two growth-suppressing pathways downstream of Ft.
(A) Protein model of the intracellular domain of Fat showing the transmembrane domain (TM), regions conserved with mammalian Fat4 (blue, A–F) (defined by Pan et al., 2013), regions associated with the major growth suppressive function of Fat (red) (HM, Bossuyt et al., 2013; Hippo-N, Hippo-C, Matakatsu and Blair, 2012; H2, Zhao et al., 2013), region required for Dco binding (green) (Sopko et al., 2009), mutV region (orange) (Pan et al., 2013), Su(DN) region (purple) (Matakatsu and Blair, 2012), and two point mutations, ftsum (Bossuyt et al., 2013) and ft61 (this study). Size and position of regions are drawn to scale relative to the ICD. (B–C) Mosaic adult eye assay. Heterozygous wild-type cells have red pigment and homozygous mutant cells lack pigment. Compared to (B) control FRT40A mosaic eyes, (C) ft61 mosaic eyes are larger and have more mutant tissue. (D) ftGrv/ft8; ft+ adult wing and (D′–D‴) confocal slice of a wing disc showing that Fbxl7 (red) is localized to the apical membrane similar to E-cad (green). (E) ftGrv/ft8; ftΔD adult wing and (E′–E‴) confocal slice showing that Fbxl7 (red) apical localization is disrupted. (F) ftGrv/ft8; ftΔF adult wing and (F′–F‴) confocal slice showing that Fbxl7 (red) apical localization is normal and similar to that in D′–D‴. (G) Confocal slice of a disc containing a MARCM ft61 clone (green) and stained for Fbxl7 (red) and E-cad (blue). Fbxl7 apical localization is normal in ft61 cells (H) Co-immunoprecipitation experiment in S2 cells. Fat-V5, Fat61-V5, and FatΔF-V5 pull down with FLAG-Fbxl7, whereas pull down of FatΔD-V5 and FatmutV-V5 is reduced. Expressed Fat proteins contain only transmembrane and cytoplasmic regions (ICD). (I–M) Wing imaginal discs (and associated leg and haltere discs) at low magnification. Compared to (I) control tub-Gal4 discs, (J) ftGrv/ft8 and (K) ft61/ft8 discs are larger and have more folds. (L) Ubiquitous expression of Fbxl7 does not rescue ftGrv/ft8 disc overgrowth. (M) Ubiquitous expression of Fbxl7 rescues disc overgrowth of ft61/ft8. (N–O) Adult wing from (N) control tub-Gal4 and (O) ubiquitous expression of FLAG-Fbxl7 in an ft61/ft8 background. Animal lethality is rescued. (P–Q′) Confocal slice through the eye imaginal disc showing MARCM clones (green) and anti-Diap1 staining (red). (P–P′) dco3 clones have elevated Diap1 levels and are overgrown, whereas (Q–Q′) dco3 clones expressing FLAG-Fbxl7 have wild-type Diap1 levels and are reduced in size.
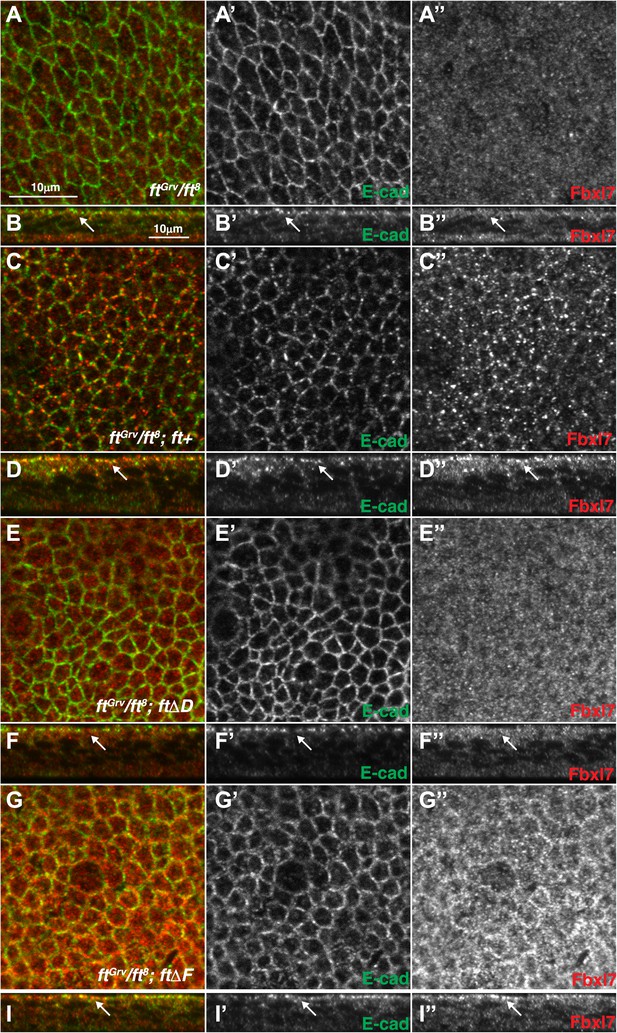
Additional images of Fbxl7 localization in Fat deletion backgrounds.
Confocal sections of wing disc cells stained with anti-Fbxl7 (red) and anti-E-cad (green) in different ft genetic backgrounds. (A–A″, C–C″, E–E″, G–G″) show apical sections and (B–B″, D–D″, F–F″, H–H″) show z-sections. (A–B″) ftGrv/ft8, (C–D″) ftGrv/ft8; ft+, (E–F″) ftGrv/ft8; ftΔD, (G–H″) ftGrv/ft8; ftΔF. Arrows indicate location of apical membrane. Fbxl7 apical localization is lost in ftGrv/ft8; ftΔD.
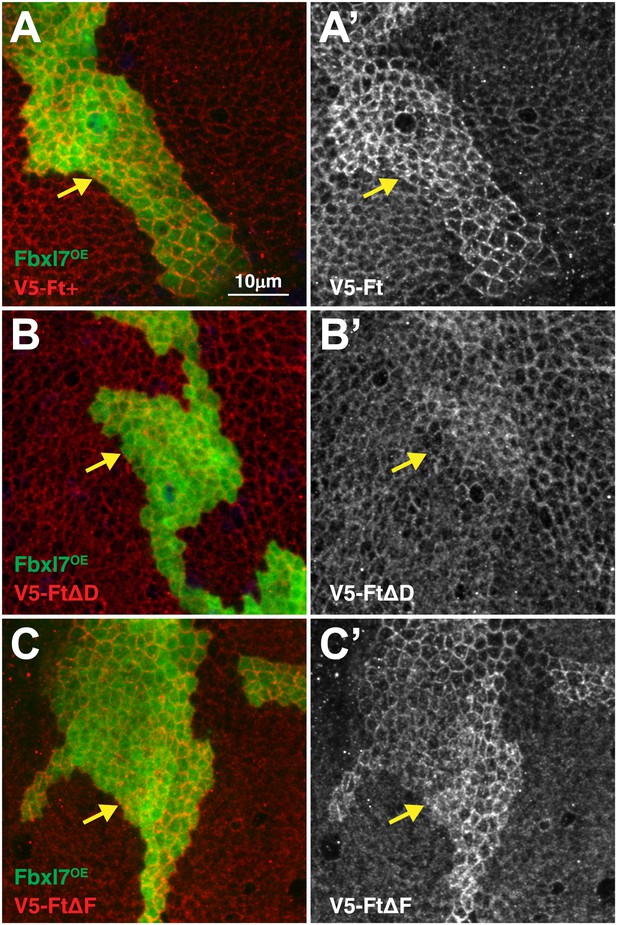
Domain D of Ft is required for the effects of Fbxl7 on Ft localization.
Confocal projection of apical membrane in wing disc cells stained for anti-V5 (red) with clones overexpressing FLAG-Fbxl7 (green, marked by GFP) in different genetic backgrounds. Ft proteins are N-terminally tagged with V5. (A–A′) V5-Ft+, (B–B′) V5-FtΔD, (C–C′) V5-FtΔF. Arrows indicate edge of clones. Apical levels of V5-FtΔD protein are not increased upon Fbxl7 overexpression.
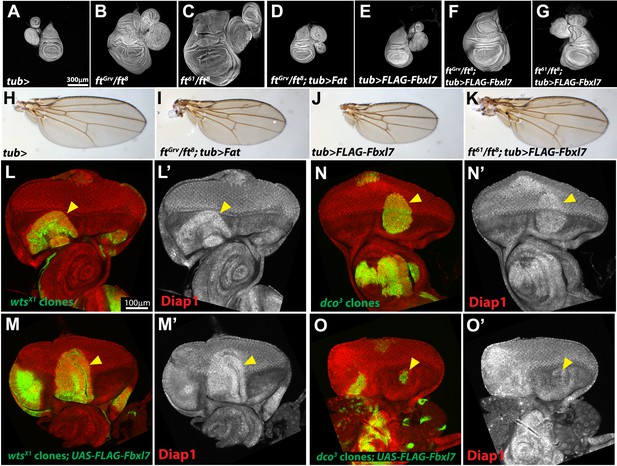
Additional characterization of Fbxl7 rescue experiments.
(A–G) Wing imaginal discs (and associated leg and haltere discs) at low magnification. Expanded set of panels compared to Figure I–M. Compared to (A) control tub-Gal4 discs, (B) ftGrv/ft8 and (C) ft61/ft8 discs are larger and have more folds. (D) Ubiquitous expression of Fat rescues ftGrv/ft8 disc overgrowth. (E) Ubiquitious expression of Fbxl7 does not obviously change disc size from wild type. (F) Ubiquitous expression of Fbxl7 does not rescue ftGrv/ft8 disc overgrowth. (G) Ubiquitous expression of Fbxl7 rescues disc overgrowth of ft61/ft8. (H–K) Adult wings. (H) Control adult tub-Gal4 wing. (I) Ubiquitous expression of Fat can rescue ftGrv ft8 animal lethality. (J) Ubiquitous expression of Fbxl7 does not affect viability and wings are smaller. (K) Ubiquitous expression of Fbxl7 rescues animal lethality of ft61/ft8. (L–O′) Confocal slice through the eye imaginal disc showing MARCM clones (green) (arrowheads) and anti-Diap1 staining (red). (L–L′) wtsX1 clones have elevated Diap1 levels and are overgrown. (M–M′) Expressing FLAG-Fbxl7 in wtsX1 clones does not effect the elevated Diap1 levels or overgrowth. (N–N′) dco3 clones have elevated Diap1 levels and are overgrown, whereas (O–O′) dco3 clones expressing FLAG-Fbxl7 have wild-type Diap1 levels are reduced in size.
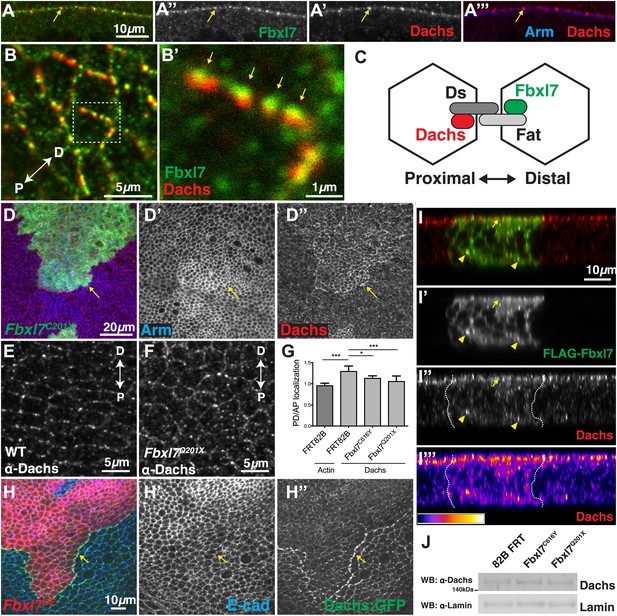
Fbxl7 regulates the localization of Dachs.
(A–A″) Confocal slice through a bend in the wing disc showing (A′) Dachs (red) and (A″) Fbxl7 (green) localize at subapical membrane. (A‴) Like Fbxl7, Dachs is apical to the adherens junction marked by Arm (blue). (B–B′) Confocal slice through the apical surface of the wing disc, specifically the dorsal edge of the pouch, showing Dachs (red) and Fbxl7 (green) staining. Dachs and Fbxl7 puncta abut each other on either side of the cell boundary. Proximodistal axis indicated as P<−>D. (C) Diagram of polarized wing disc cells in which Dachs is enriched on the distal side and Fbxl7 is on the proximal side, linked by their association to Dachsous and Fat, respectively, which bind across cells. (D–D″) Apical confocal slice of MARCM Fbxl7Q201X clones (green) and staining for Dachs (red) and Arm (blue). Dachs levels are elevated in clones. (E–G) Apical confocal slice with staining for Dachs in (E) wild-type or (F) Fbxl7Q201X discs. Images are from the dorsal edge of the pouch and are aligned so the proximodistal axis is vertical. Dachs enrichment on P/D membrane, seen in (E) wild-type discs, is impaired in (F) Fbxl7Q201X discs. (G) Quantification of Dachs P/D enrichment in wing discs. Dachs is localized in a P/D direction, whereas Actin is not. Dachs P/D asymmetry is impaired in both Fbxl7C616Y and Fbxl7Q201X discs. Significance calculated with one-way ANOVA test. ***p ≤ 0.001, *p ≤ 0.05. Error bars indicate SD. (H–H″) Apical confocal slice of FLAG-Fbxl7 overexpressing clones (red, cells marked by RFP) and staining for anti-GFP (green, Dachs:GFP) and E-cad (blue). Apical Dachs levels within the clone are reduced, and Dachs is enriched at the edge of the clone. (I–I‴) Confocal z-section of a wing disc with a FLAG-Fbxl7 expressing clone (green) and stained for Dachs (red). FLAG-Fbxl7 and Dachs co-localize to apical membrane (arrow) and intracellular puncta (arrowheads). (I″–I‴’) Cytoplasmic levels of Dachs are slightly elevated within the clone. (I‴) Heat map of I″. (J) Western blots from wing disc lysates. Endogenous Dachs protein levels are not changed in Fbxl7 mutant wing discs compared to control.
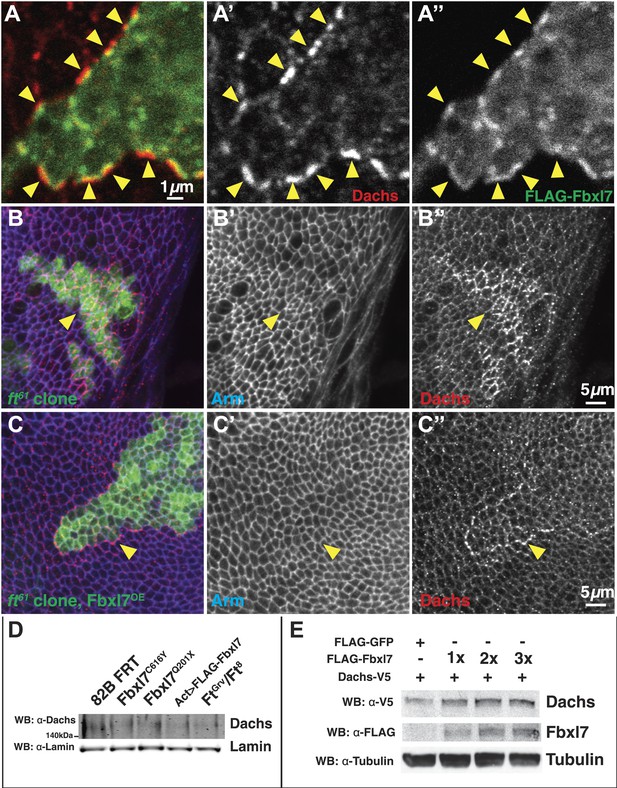
Additional Dachs tissue staining and Dachs levels in wing discs and S2 cells.
(A–A′) Confocal slice through apical membrane of a clone expressing FLAG-Fbxl7 (green) in the wing disc and staining for Dachs (red). In wild-type cells bordering the clone, Dachs is enriched at the boundary. Puncta of Dachs in wild-type cells are apposed across the boundary with puncta of FLAG-Fbxl7 in overexpressing clones (arrowheads). (B–C) Confocal slice through apical membrane of ft61 clones (green, marked by GFP) and stained for anti-Dachs (red) and anti-Arm (blue). Arrowheads indicate edge of clone. (B) Dachs levels are higher in ft61 clones. (C) Overexpression of FLAG-Fbxl7 in ft61 clones reduces apical Dachs levels, to levels comparable to neighboring wild-type cells. Dachs levels are enriched at the edges of the clone. (D) Additional western of endogenous Dachs protein in dissected wing discs. Dachs protein levels do not change in Fbxl7 mutant, Fbxl7 overexpressing, or ft mutant backgrounds compared to control. (E) Western analysis of transfected Dachs-V5 protein levels in S2 cells. Transfecting increasing doses of FLAG-Fbxl7 does not change Dach-V5 levels.
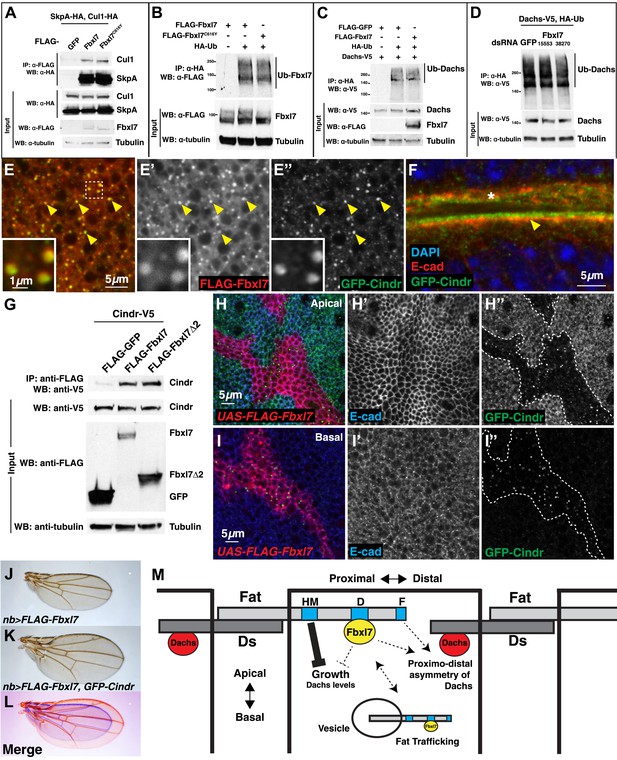
Fbxl7 does not affect Dachs ubiquitylation, and Fbxl7 affects the localization of Cindr.
(A) Co-immunoprecipitation assay from S2 cells. SkpA-HA and Cul1-HA immunoprecipitates with FLAG-Fbxl7. (B) In-vivo Fbxl7 ubiquitylation assay in S2 cells. FLAG-Fbxl7 and FLAG-Fbxl7C616Y are ubiquitylated in vivo. (C–D) In-vivo Dachs ubiquitylation assay in S2 cells. Dachs-V5 is ubiquitylated under wild-type conditions, and does not change with (C) overexpression of FLAG-Fbxl7 or (D) knockdown of Fbxl7 with two different dsRNAs. (E–E″) Confocal slice showing localization of FLAG-Fbxl7 (red) and GFP-Cindr (green) in puncta (arrowheads). (F) Confocal slice through a bend in the wing disc. GFP-Cindr (green) localizes to subapical membrane, apical to E-cad (red). Asterisk indicates an adjacent bend in the tissue. (G) Co-immunoprecipitation experiment in S2 cells. Cindr-V5 pulls down with full length FLAG-Fbxl7, and FLAG-Fbxl7Δ2, which contains only the LRR domains. (H–I″) Confocal slice in a disc with clones overexpressing FLAG-Fbxl7 (red, cells marked by myr-RFP) in a GFP-Cindr background. (H–H″) An apical plane shows loss of apical GFP-Cindr within the clone, and (I–I″) a basal plane shows accumulation of GFP-Cindr in puncta. (J) Compared to (J) nb>FLAG-Fbxl7 alone, (K) overexpressing GFP-Cindr partially rescues the small wing phenotype. (L) Merge shows J blue and K in red. (M) Model of Fbxl7 as a component of Fat signaling. Not drawn to scale.
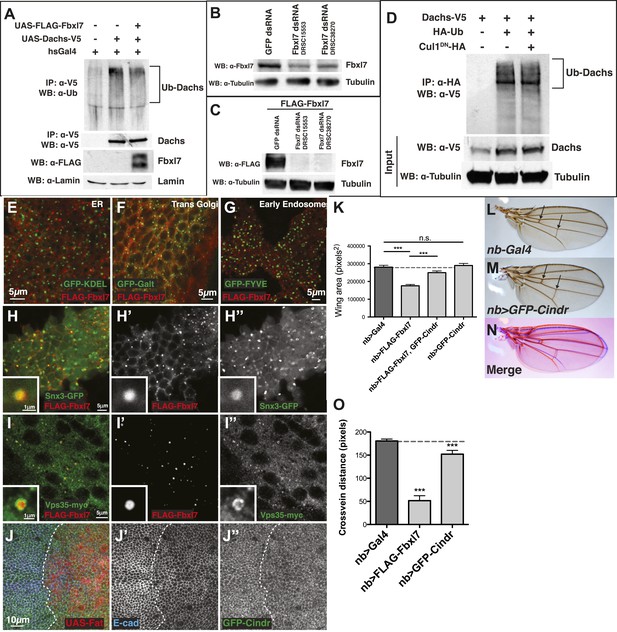
Additional analysis of Dachs ubiquitylation, vesicle markers, and Cindr.
(A) In-vivo ubiquitylation assay of Dachs-V5 from imaginal discs. Expressing FLAG-Fbxl7 does not alter ubiquitylation of Dachs-V5. (B) Knockdown of endogenous Fbxl7 and (C) knockdown of transfected FLAG-Fbxl7 by two different dsRNAs (DRSC15553, DRSC38270) in S2 cells. (D) In-vivo ubiquitylation assay of Dachs-V5 from S2 cells. Expressing Cul1DN does not affect Dachs-V5 ubiquitylation. (E–J″) Confocal slice showing localization of FLAG-Fbxl7 (red) and a vesicle marker (green) in the wing disc. Fbxl7 does not colocalize with a marker of the (E) ER (GFP-KDEL), (F) trans-Golgi (GFP-Galt), or (G) early endosomes (GFP-FYVE). (H–I″) Fbxl7 partially colocalizes with (H–H″) Snx3-GFP and (I–I″) Vps35-myc in vesicles. Insets shows magnification of vesicles with overlap. (J–J″) Confocal section through the apical surface of a wing disc expressing UAS-Fat (marked with mRFP) in the posterior compartment (engrailed-Gal4) in a GFP-Cindr (green) background and staining for E-cadherin (blue). GFP-Cindr apical localization is unchanged with Fat overexpression. (K) Genetic interaction of Fbxl7 and Cindr experiment and quantification of adult wing size from expression of genes in the wing pouch (nb-Gal4). UAS-GFP-Cindr overexpression partially rescues the small wing due to UAS-FLAG-Fbxl7 overexpression. UAS-GFP-Cindr expressing wings are wild type in size. (L–M) Compared to (L) control nb-Gal4 wings, (M) overexpressing GFP-Cindr causes wings to be rounder and crossveins closer (arrows). (O) Quantification of reduced crossveins of GFP-Cindr overexpressing wings. (K and O) Significance calculated with 1way ANOVA followed by Tukey's test. n ≥ 10 wings. ***p ≤ 0.001. Error bars indicate SD.
Additional files
-
Supplementary file 1
Localization of FLAG-Fbxl7 and vesicle markers. List of tagged proteins that localize to vesicles, are related to vesicle trafficking, and proteins interacting with Fat. Colocalization was assessed with FLAG-Fbxl7 in the wing imaginal disc.
- https://doi.org/10.7554/eLife.03383.019
-
Supplementary file 2
Sequence of oligonucleotides used.
- https://doi.org/10.7554/eLife.03383.020





