Laser ablation of Dbx1 neurons in the pre-Bötzinger complex stops inspiratory rhythm and impairs output in neonatal mice
Figures
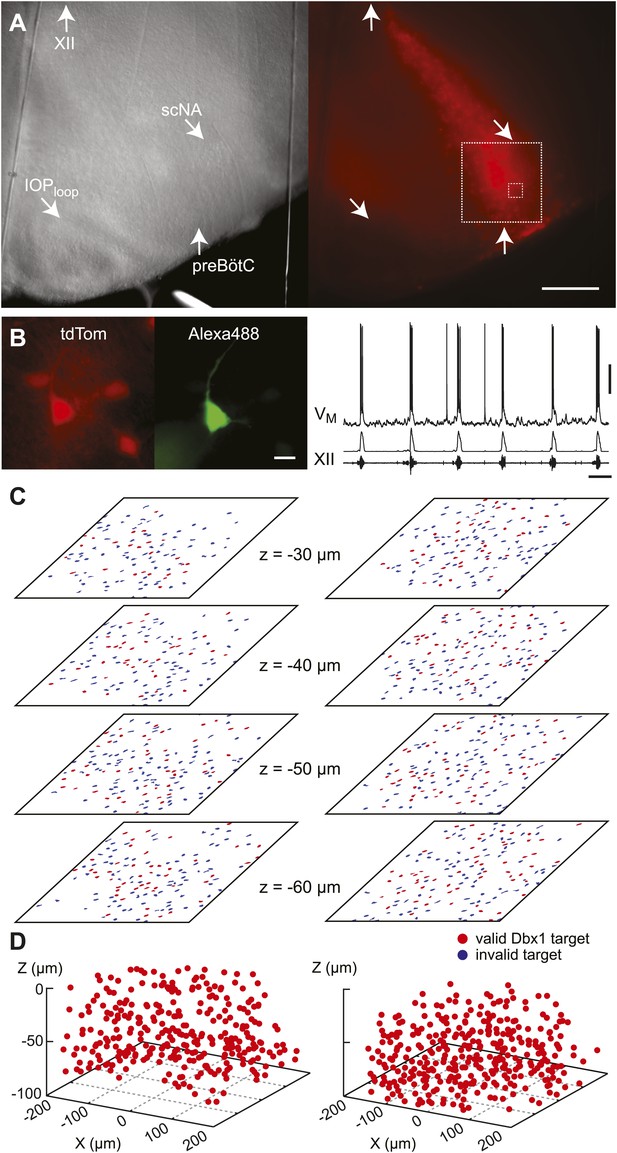
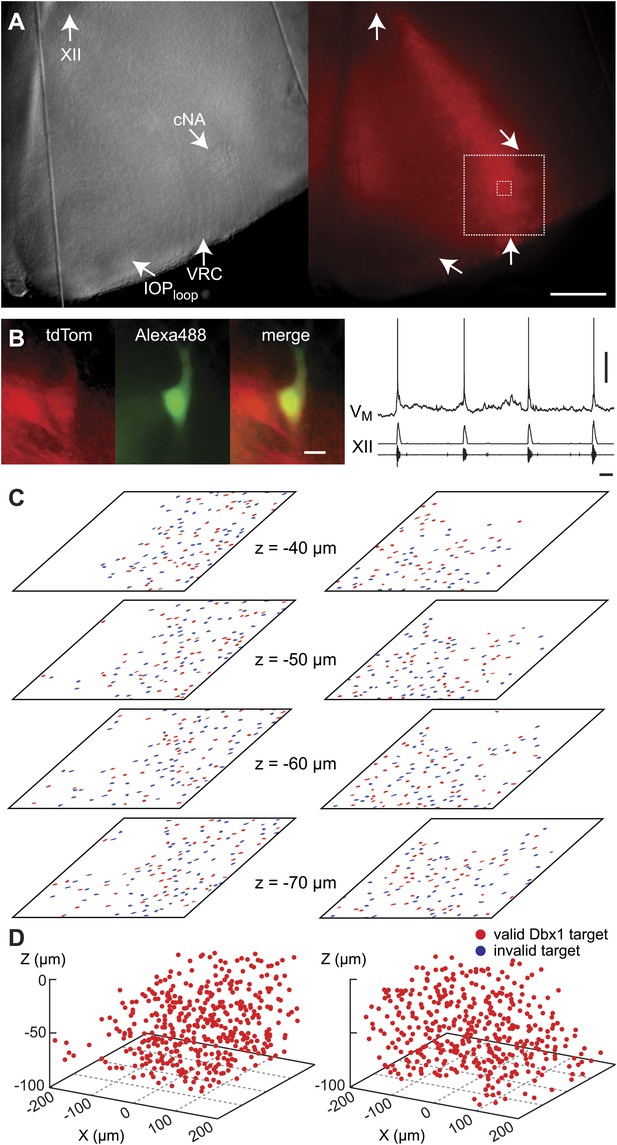
Dbx1 preBötC neurons.
(A) Bright field (left) and fluorescence (right) images of the right half of a preBötC-surface slice preparation. Anatomical landmarks are illustrated including: XII, the hypoglossal motor nucleus; scNA, semi-compact nucleus ambiguus; IOPloop, the dorsal loop of the principal inferior olive; and the ventral border of the preBötC, which is orthogonal to the IOPloop. Scale bar is 300 µm. At right, the larger white box shows the detection and ablation domain. (B) Expansion of smaller white box in A, showing tdTomato expression in Dbx1 neurons and intracellular dialysis via patch pipette with Alexa 488 from the recorded neuron whose robust inspiratory discharge is illustrated at right (scale bar is 10 µm). Respiratory motor output from the XII nerve is shown in raw and RMS-smoothed form. Voltage and time calibration bars represent 20 mV and 2 s. Baseline membrane potential in the recorded neuron was −60 mV. (C) Mask of targets showing validated Dbx1 interneuron targets (red) and regions of fluorescence that do not pass muster and were rejected as targets (blue) for focal planes at depths z = (30–60 µm). The region shown in each case maps to the 412 × 412 µm2 square shown by the larger white box in A (right). Only a subset of the masks are shown for economy of display. (D) 3D reconstruction of detected targets for all focal planes z = (10–80 µm) from the left and right preBötC. Each Dbx1 neuron is represented by a single red point centered on its soma.
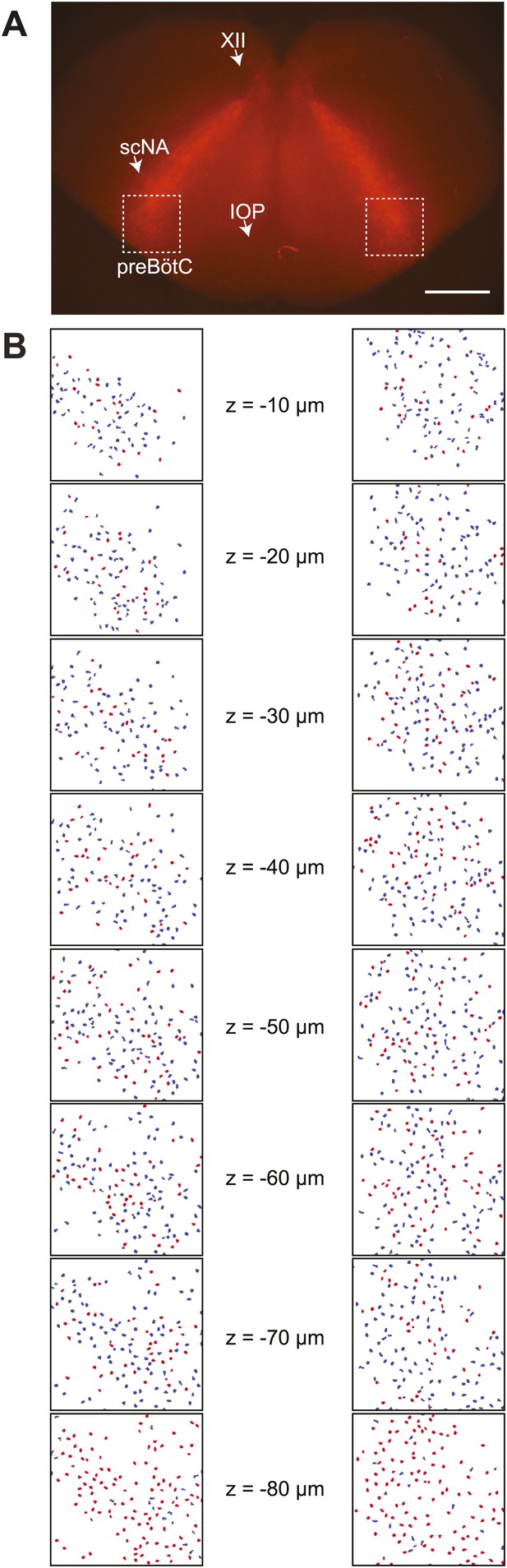
Detection of Dbx1 preBötC neurons.
(A) Fluorescent image of a transverse slice from a Dbx1+/CreERT2; Rosa26tdTomato mouse pup. Anatomical landmarks are illustrated including: XII, the hypoglossal motor nucleus; scNA, semi-compact nucleus ambiguus; and IOP, the principal inferior olive. The domain for detection and ablation is indicated by the white boxes, bilaterally. Scale bar is 500 µm. (B) Mask of targets showing validated Dbx1 (red) and invalidated (blue) cells for all focal planes to a depth of −80 µm. Each image is 412 × 412 µm2 (as in Figure 1C). Image processing routines for detecting and validating Dbx1 neuron targets are detailed in ‘Materials and methods’, Figure 1—figure supplement 2, and a methodological paper (Wang et al., 2013). Note that the highest fraction of validated Dbx1 target cells is found at deeper focal planes, e.g., −80 µm due to the ‘priority rule’, which applies to overlapping ROIs in adjacent focal planes. According to the priority rule, the ROI from the deeper focal is accepted as a ‘bona fide’ target and the redundant ROI at the superficial level is rejected. Also see Figure 1—figure supplement 2C,D.
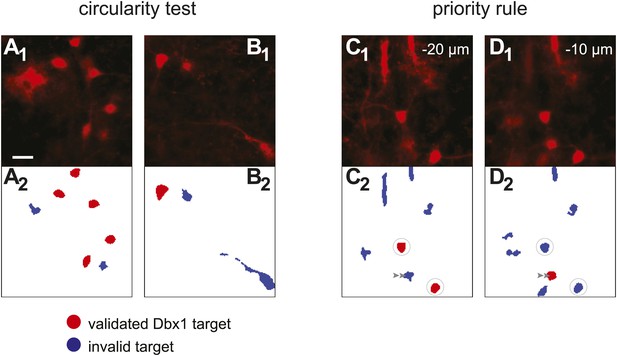
Detection of Dbx1 neuron targets via fluorescence and image processing.
(A1, B1, C1, D1) Images from the preBötC of Dbx1+/CreERT2; Rosa26tdTomato mice showing tdTomato in neurons derived from Dbx1-expressing precursors (i.e., Dbx1 neurons). Scale bar in A1 is 20 µm and applies to all panels. C1 and D1 show the same field of view at two different depths (−20 and −10 µm, respectively). (A2, B2, C2, D2) Masks of ROIs obtained by analyzing the corresponding images above. Red ROIs are deemed valid targets by the circularity test, which evaluates somatic shape; blue ROIs that fail the circularity test are rejected. Circularity analyses distinguish somata from auto-fluorescent detritus (A1, A2) as well as contiguous soma-dendrite images (B1, B2) and isolated segments (shafts) of dendrites (C1, C2, D1, D2). Non-somatic auto-fluorescence is rejected because it does not accurately indicate underlying neurons. Dendritic segments are not valid targets because they are difficult to target in the ablation phase of the experiments and their cell bodies are detectable in adjacent focal planes. Often, a cell rejected by the circularity test in one focal plane (e.g., C2, graygray double arrowhead) is validated in the adjacent plane (D2, graygray double arrowhead). When ROIs that pass the circularity test are detected in more than one focal plane, they are validated or rejected according to the priority rule. ROIs from a deeper focal plane (−20 µm) are validated by circularity and thus colored red (C2, circled ROIs). Subsequent detection of overlaying ROIs at the superficial focal plane (−10 µm), which also pass the circularity test, are nonetheless rejected by the priority rule and thus colored blue (D2, circled ROIs). These criteria for target detection are more fully described in ‘Materials and methods’ and Wang et al. (2013).

Average number of Dbx1 neurons detected at each acquisition depth from z = 0 (surface) to z = −80 µm in preBötC-surface slices and control slices with the ventral respiratory column (VRC) exposed at the slice surface.
The number of Dbx1 neurons detected per focal plane per side (in 10-µm increments of the focal plane) is shown individually for each individual experiment (graygray unfilled circles) along with the mean ±SD for all experiments (black unfilled squares with black lines showing SD).
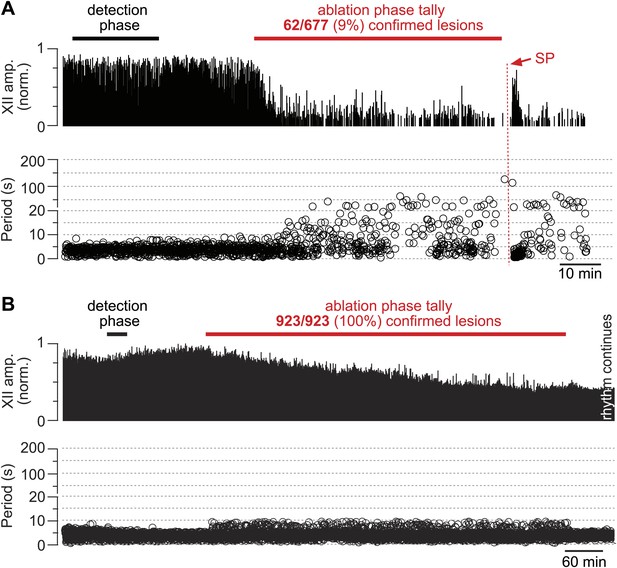
Cumulative serial ablation of Dbx1 neurons in preBötC-surface slices (A) and control slices whose surface exposes the ventral respiratory column, not preBötC (B).
(A and B) The x-axis is a timeline. The y-axis plots XII amplitude (normalized units, top) and respiratory period (bottom). The respiratory period axis is continuous (0–200 s) but plotted with two scales. Major ticks are separated by 10 s from 0 to 20 s (with unlabeled minor ticks at 5 s increments), and thereafter major ticks are plotted in 100 s divisions from 21 to 200 s (with unlabeled minor ticks at 50 s increments). The discontinuity in the y-axis stops at 20 s (lower portion) and starts at 21 s (upper portion). There is one data point for every individual respiratory period measured. The recording in A is no longer displayed after 6 min of XII quiescence. Substance P (SP) injection in A is displayed at higher sweep speed in Figure 5C. The recording in B is no longer displayed after 90 min of continuous stable XII output following the end of the ablation phase. Time calibrations in A and B are shown separately.

Cellular laser ablation and confirmation.
(A) The image acquired during maximum-intensity Ti:sapphire laser scanning of the target cell with a 560–615 nm band-pass filter, which indicates cell destruction. This image was acquired with higher digital magnification compared to panels B–D; scale bar is 2 µm. (B1–2) Images of native tdTomato expression in Dbx1 preBötC neurons before (B1) and after (B2) a single cell laser ablation. The target cell (arrowhead) is visible pre-lesion but not in the post-lesion image. Neighboring (unlesioned) neurons are unaffected. Scale bar of 10 µm applies to all images in B–D. (C) Bright field images of the target cell (arrowhead) prior to laser lesion. (D1–5) Images of the target cell post-lesion (arrowhead) at 5-µm increments in the z plane. The focal plane in C was normalized to z = 0 µm for relative comparison with panels D1–5.
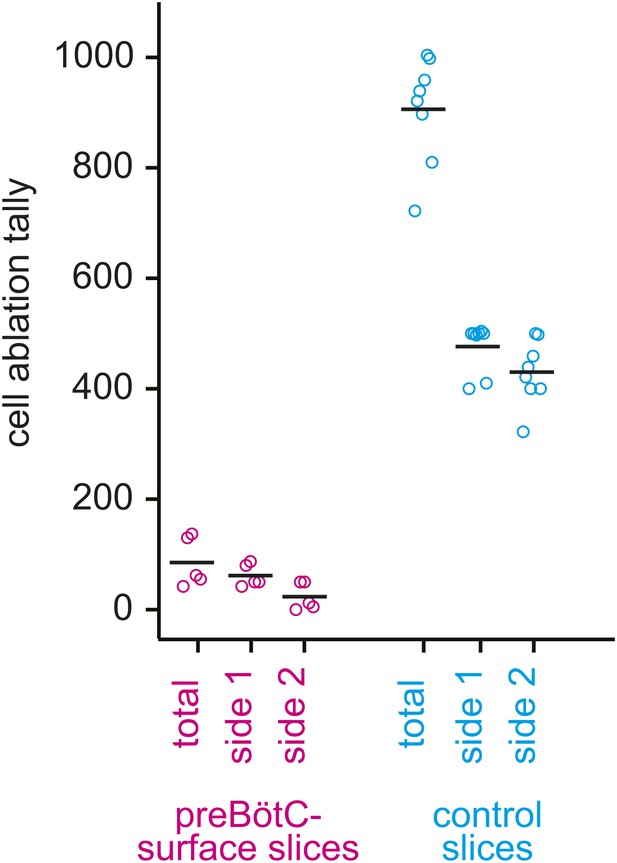
Cumulative tally of laser ablations for preBötC-surface slices (magenta) and control slices whose surface exposes the ventral respiratory column, not preBötC (cyan).
The total tally and the individual side tallies are shown for each preparation. Black bars show the mean. For preBötC-surface slices, the tally was always lower on the side that was being lesioned when the rhythm stopped because rhythm cessation halted the ablation sequence.

Ablation effects on respiratory frequency and the amplitude of XII motor output.
(A–D) Measurements are displayed in light grey and red for preBötC-surface slices and dark grey and blue for control slices that expose the ventral respiratory column (VRC). (A) XII amplitude and (B) respiratory frequency for preBötC-surface and control slices are plotted vs cumulative percent of total lesions during the ablation phase (bars show SD). Inset in B shows respiratory period in lieu of frequency (bars show SD) for preBötC-surface slices. (C and D) The regularity score (RS) is plotted vs cumulative percent of total lesions for preBötC-surface (C) and control slices (D). B, C, and D are plotted on semi-log axes. B and C are labeled with subordinate ticks at 2, 4, and 6. Tick labels are omitted from D because they match C exactly. B (inset) has linear axes.
Dbx1 neurons in the ventral respiratory column.
(A) Bright field (left) and fluorescence (right) images of the right half of a control slice preparation. Anatomical landmarks are illustrated including: XII, the hypoglossal motor nucleus; cNA, the compact division of the nucleus ambiguus; IOPloop, the ventral portion (loop) of principal sub-nucleus of the inferior olive; and VRC, the ventral border of the ventral respiratory column. Scale bar is 300 µm. At right, the larger white box shows the detection and ablation domain. (B) Expansion of smaller white box in A, showing tdTomato expression in Dbx1 ventral respiratory column neurons (scale bar is 10 µm), one of which was recorded. Intracellular dialysis via patch pipette with Alexa 488 is visible in the recorded neuron whose inspiratory depolarization and discharge pattern are illustrated at right. Respiratory motor output from the XII nerve is shown in raw and RMS-smoothed form. Voltage and time calibration bars represent 20 mV and 1 s. (C) Masks of targets showing validated Dbx1 interneuron targets (red) and regions of fluorescence that do not pass muster and were rejected as targets (blue) for focal planes at depths z = (40–70 µm). (D) 3D reconstruction of detected targets for all focal planes z = (0–80 µm) in the ventral respiratory column (VRC) of the left and right side. A single red point centered on its soma represents each Dbx1 neuron. The highest fraction of accepted Dbx1 target cells is found at deeper focal planes (see Figure 1—figure supplement 2 and ‘priority rule’ explained in ‘Materials and methods’).
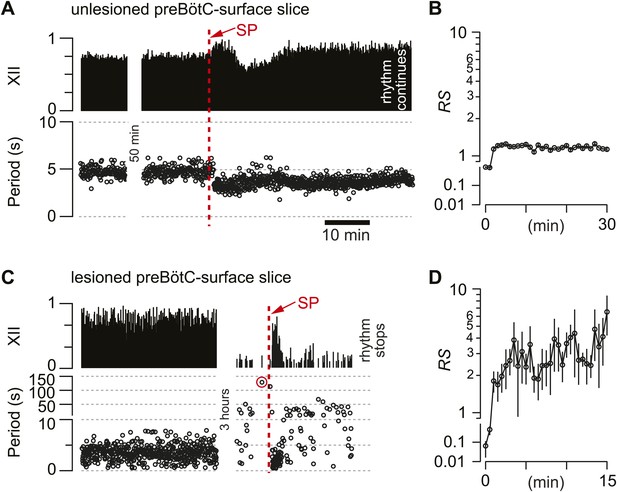
Substance-P (SP) injections in preBötC-surface slices.
(A) SP bolus injected in an un-lesioned preBötC-surface slice. XII output magnitude is plotted with cycle period as a time series. (B) Semi-log plot of regularity score (RS) for 30 min after SP injection from the slice preparation in A. RS axis is continuous but plotted with two scales. (C) preBötC-surface slice shown in the acquisition phase (left) and during the ablation phase (right), which were separated by a time gap of 3 hr. After 120 s of quiescence (data point circled in red), SP injection revived the rhythm transiently. (D) Semi-log plot of RS for 15 min after SP injection from the slice preparation in C. Data in C and D were from the same preparation as in Figure 2A.
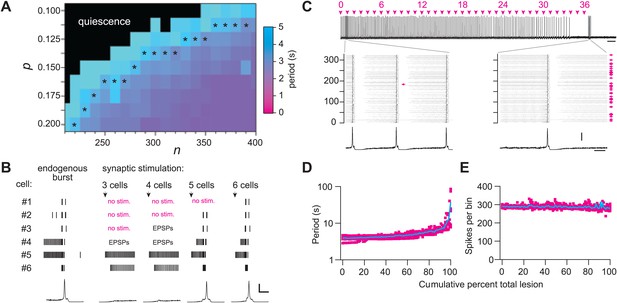
Numerical simulations.
(A) Networks of Dbx1 preBötC neurons with population size (n) and synaptic connection probability (p). Blocks show the mean cycle period according to the colorimetric scale (right) for 10 (or more) realizations of the network for each (n,p) pair. Asterisks denote networks that generated respiratory-like (∼4 s) cycle periods in ≥80% of individual realizations of the network. (B) Focal glutamatergic stimulation of constituent neurons in a model network (n,p) = (330,0.125). Network-wide bursts can be evoked when five or more individual cells are stimulated. These simulations mimic holographic laser-mediated glutamate un-caging experiments (Kam et al., 2013b) and are included because they bolster confidence that our model networks accurately capture features and behaviors of the preBötC in newborn mice. Raster plots show spike activity in six constituent neurons randomly selected from the network and focally stimulated (see ‘Materials and methods’ for numerical simulation of glutamate un-caging protocol). If focal stimulation evoked EPSPs (not spikes) then the raster reports ‘EPSPs’; spikes are indicated by short vertical lines. From left to right, the number of stimulated units increments by one; five (or more) units evoked an inspiratory-like burst. A running-time histogram of network activity is shown at the bottom. Calibration bars represent 100 spikes/10-ms bin (vertical) and 0.5 s (horizontal). (C) Running-time histogram for one simulation of sequential ablation in a network (n,p) = (330, 0.125). Cell ablation tally is shown (top). Time calibration is 30 s. Spikes-per-bin calibration bar is the same as the inset (lower), 100 spikes/10-ms bin. Insets show a raster plot of spike activity in the entire network with a running-time histogram. The numerical y-axis reports cell index for each neuron model in the network. Left inset shows the first ablation (magenta arrow). Right inset shows all cumulative 36 ablations (magenta arrows). Time calibration for both insets is 1 s (at right). (D) Cycle period and (E) spikes-per bin (i.e., a measure of the magnitude of simulated network output as in C) are plotted vs cumulative percent of total ablations for 10 networks with (n,p) = (330, 0.125). D plotted in semi-log axes, E in linear axes. Magenta shows data from individual networks, cyan plots the mean response.
Numerical simulations of Dbx1 neuron laser ablation experiments.
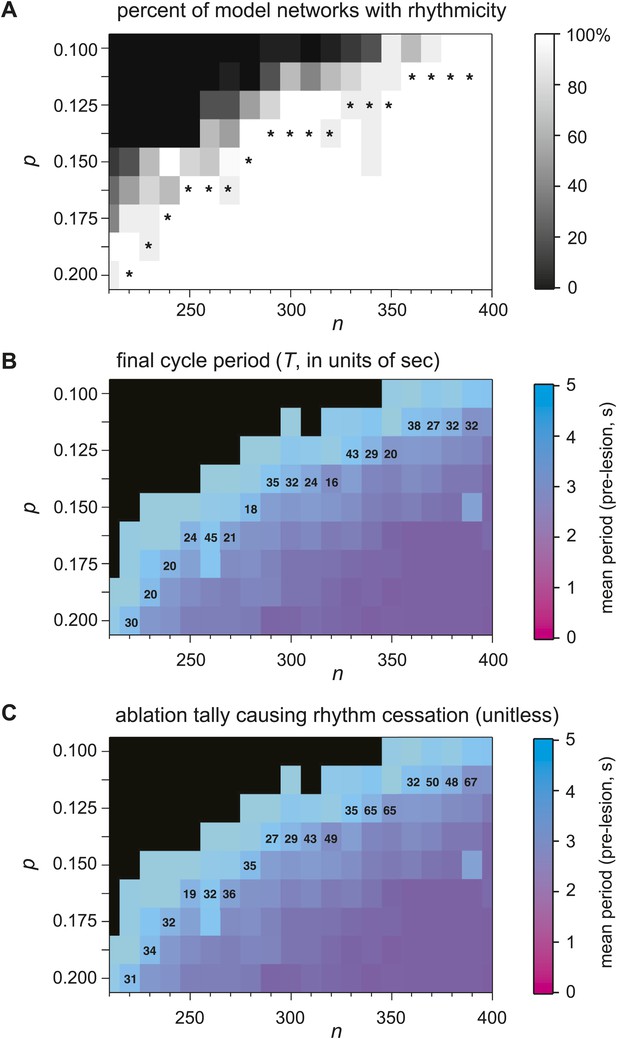
Networks of Dbx1 preBötC neurons parameterized by population size (n) and synaptic connection probability (p). Erdős-Rényi random directed graphs G(n,p) (Newman et al., 2006) determined the underlying connectivity structure. Each node in G(n,p) was populated by a Rubin-Hayes preBötC neuron model (Rubin et al., 2009) with dynamic excitatory synapses for links. Each block in the panels reports a measure of network performance. (A) The grey scale reports the percent of model networks that generated spontaneous rhythmic activity. Asterisks denote networks that generated respiratory-like cycle periods in ≥80% of individual realizations, which were then subjected to simulated laser ablation experiments (results in B and C). (B) The colorimetric scale reports the mean cycle period for 10 (or more) realizations of the network for each (n,p) pair (same as Figure 6A and panel C). Networks with asterisks (from Figure 6A and this figure's panel A) were subject to laser ablations in random sequence; the numbers in the blocks report the average final cycle period (in s) prior to rhythm cessation in the lesioned network at each (n,p) pair. (C) The numbers in the blocks report the average cell ablation tally at the point of rhythm cessation for five or more laser ablation simulations.

Dbx1 preBötC neurons with premotor function.
(A) Fluorescence and (B) bright field images of a slice preparation. Anatomical landmarks are illustrated including: XII, the hypoglossal motor nucleus; scNA, semi-compact division of the nucleus ambiguus; IOPloop, the ventral loop of the principal inferior olive, and the ventral surface of the preBötC. Scale bar is 100 µm and applies to A and B. A patch-recording pipette is visible, marking the inspiratory-modulated neuron detailed in C–E. A dotted circle indicates the tip of the pipette and cell body. (C and D) tdTomato expression, intracellular dialysis of Alexa 488, and merged image (C) from the inspiratory neuron shown with XII nerve output (D). Voltage and time calibration bars represent 20 mV and 1 s. Baseline membrane potential in the recorded neuron was −60 mV. (E) Antidromic activation of the Dbx1 inspiratory neuron from C and D. Action potentials were evoked by XII stimulation (left) and intracellular 5-ms supra-threshold current pulses (middle). When the antidromic XII stimulus was preceded immediately by a supra-threshold intracellular current pulse, the antidromic spike was occluded (collision test, right). Several sweeps, all from a −62 mV baseline membrane potential, are superimposed with vertical offset in each case. Voltage calibration is the same as panel D. Applied current (Iapp) calibration is shown. Time calibration bar for E is 25 ms.
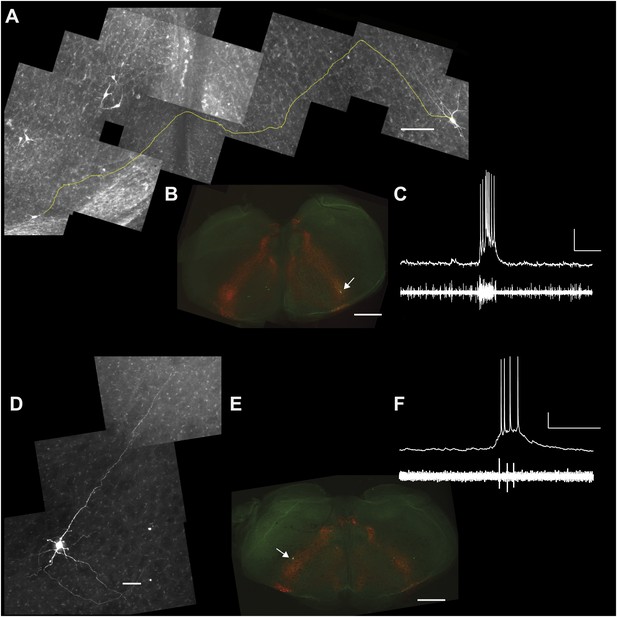
Commissural and premotor projections of inspiratory Dbx1 preBötC neurons.
(A) Biocytin-filled and reconstructed Dbx1 preBötC neuron with commissural axon projection. The axon, which meanders in depth in this confocal image stack, was digitally traced (yellow) and superimposed in one plane for display. Axon trajectory crosses the midline of the slice and enters the preBötC contralaterally. Scale bar is 25 µm. (B) Mosaic image of the entire slice. The biocytin-filled soma (green) of neuron in A is shown at lower right (white arrow). Scale bar is 200 µm. Panels A and B have exactly the same orientation (dorsal up, ventral down). (C) Inspiratory discharge from the neuron in A and B. Top trace is membrane potential of the recorded Dbx1 preBötC neuron. Lower trace is XII output. Scale bars are 10 mV and 0.5 s. (D) Biocytin-filled and reconstructed Dbx1 preBötC neuron that projects toward the XII motor nucleus. Scale bar is 25 µm. The axon remained largely coplanar and thus is readily visible, except near its distal tip. In Figure 8—figure supplement 1, this same neuron is shown with a digitally traced (yellow) axon superimposed on the confocal image. (E) Mosaic image of the entire slice. Neuron in D is shown at lower left (white arrow). Scale bar is 200 µm. Panels D and E have exactly the same orientation (dorsal up, ventral down). (F) Inspiratory discharge from the neuron in D and E. Top trace is membrane potential of the recorded Dbx1 preBötC neuron. Lower trace is XII output. Scale bars are 10 mV and 0.5 s.
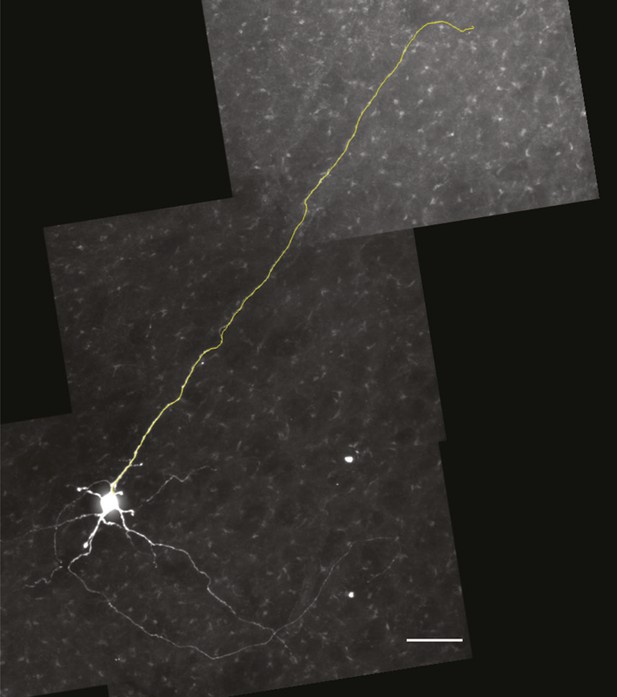
Magnified view of the Dbx1 preBötC neuron from Figure 8D-F in which the axon has been digitally traced in the confocal stack and superimposed over the image to better illustrate the axon projection toward the XII motor nucleus.
Scale bar is 25 µm.
Additional files
-
Supplementary file 1
Numerical simulations of Dbx1 neurons in model preBötC networks subjected to cumulative laser ablation experiments. Erdős-Rényi random directed graphs G(n,p) were populated with Rubin-Hayes preBötC neuron models at each node, and their links were described by excitatory synapses, as described for Figure 6 and Figure 6—figure supplement 1 above. In numerical simulations, the resulting network models with very high probability of generating rhythm and respiratory-like cycle period (∼4 s, indicated by asterisks in Figure 6A and Figure 6—figure supplement 1) were subjected to piecewise cumulative ablation protocols like slice experiments (Figures 2–4). The parameters describing the model networks (number of neurons n and synaptic connection probability p) are listed below in columns 1 and 2. Each ablation experiment was simulated five or more times. The maximum period (in s) and cumulative ablation tally (unitless) required to stop the rhythm are listed in the table for each individual realization of the network model along with average values for these characteristic measures. The networks deemed to be most representative of the preBötC, i.e., (n,p) = (320, 0.1375), (330, 0.125), (340, 0.125) were simulated 16 times each.
- https://doi.org/10.7554/eLife.03427.018
-
Supplementary file 2
Discrete network simulations. As in the main text and the table above, parameters (n,p) represent the number of constituent neurons and connection probability. Here the networks are Erdős-Rényi static directed random graphs G(n,p) (as in Figure 6 and Figure 6—supplement 1); that is, the nodes are not populated with dynamical models and the interconnections between nodes are simply static directed links (rather than dynamical synapses). Each network (static graph) below was subjected to 100 random deletions. During the ablation sequence, we computed the following global network metrics: K-core, the number of strongly connected components (SCC), and local network metrics: local cluster coefficient, closeness centrality, and betweenness centrality. The initial (first) and final (last) values for the global and local measures are plotted side by side in the appropriate columns below. We also computed the initial in- and out-degree (i.e., the average number of directed connections in and out), percentage drop in the final average in-degree, and the percentage-drop of the final average out-degree. The ‘Ave.’ row reports average change (in percent) for K-core, SCC, cluster coefficient, closeness centrality, and betweenness centrality, as well as the average in- and out-degree for initial and final states of the network. Definitions for the characteristic measures are elaborated in the ‘Materials and methods’.
- https://doi.org/10.7554/eLife.03427.019





