Allosteric signalling in the outer membrane translocation domain of PapC usher
Figures
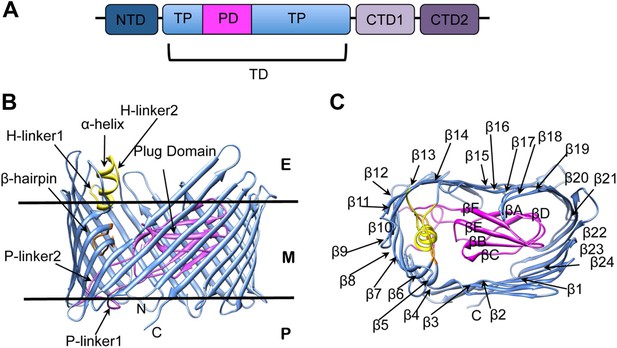
PapC usher organization and detail of its translocation domain.
(A) A diagram of the domain organization of PapC usher. NTD (dark-blue) represents the N-terminal domain, CTD1 (light-violet) and CTD2 (dark-violet) represent the C-terminal domains; TD represents the translocation domain, comprising the TP (translocation pore, light-blue) and the PD (plug domain, magenta). (B and C): Ribbon representation of the starting model of the native translocation domain (TD) of PapC with the labels ‘N’ and ‘C’ indicating the N and C termini of the translocation channel. The β-barrel, PD (including the P-linkers), β-hairpin, and α-helix (including the H-linkers) are coloured blue, magenta, orange, and yellow, respectively. The outer membrane position is represented schematically with the labels ‘E’, ’M’, and ‘P’ indicating the extracellular side, the membrane, and the periplasmic side, respectively. Side view of the TD (B) is shown with the α-helix, β-hairpin, H-linker1, H-linker2, P-linker1, P-linker2, and PD, labelled. Extracellular top view of the TD (C) is shown with the barrel β strands labelled β1 through β24 and with the PD strands labelled βA through βF. The figures were created with Chimera (Pettersen et al., 2004).
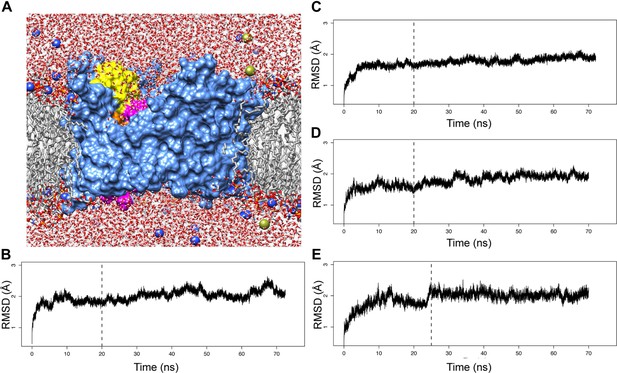
MD simulations of the native PapC TD and its mutants.
(A) Cutaway view across the membrane plane of the native PapC TD starting model in a POPE/POPG lipid bilayer (sim1, t = 0). Molecular surface of PapC TD is coloured as in Figure 1, the lipids are shown in grey with the lipid head group coloured by element, the water is coloured by element, and the ions (the Na+ in blue and the CL− in yellow) are represented as sphere. The Cα-RMSD values for each system from the starting structure (t = 0) for the native TD (B), the hairpin mutant (C), helix mutant (D), and helix-hairpin mutant (E) are plotted as a function of time.
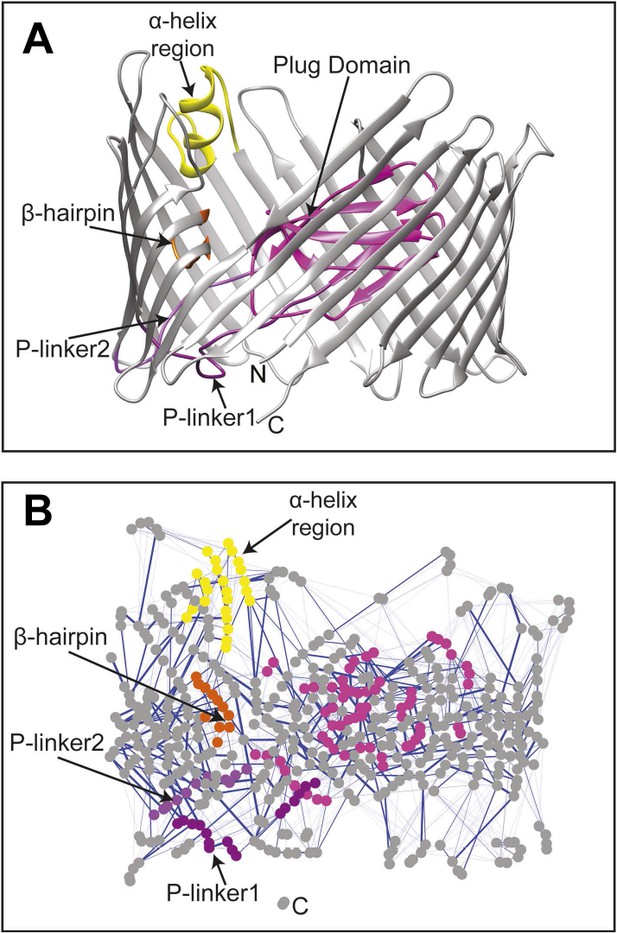
The native TD and its non-covalent interaction network (non-covalent RIN).
(A) Ribbon representation of the starting model of the native translocation domain (TD) of PapC with the labels ‘N’ and ‘C’ indicating the N and C termini of the translocation channel. The β-barrel, PD, P-linker1, P-linker2, β-hairpin, and α-helix (including the H-linkers) are coloured grey, magenta, light purple, dark purple, orange, and yellow, respectively. The α-helix, β-hairpin, P-linker1, P-linker2, and PD are labelled. (B) Non-covalent RIN representation of the native translocation domain (TD) of PapC visualized with Cytoscape 2.8.2 (Smoot et al., 2011) based on RINalyzer plug-in analysis (Doncheva et al., 2011) (see Figure 2—figure supplement 1 for the RINs of the TD mutants). The nodes (representing residues) are coloured by structural element as in (A) Edges (connecting two residues) are shown in blue, the edge width is proportional to its NCI score from lower to higher values.
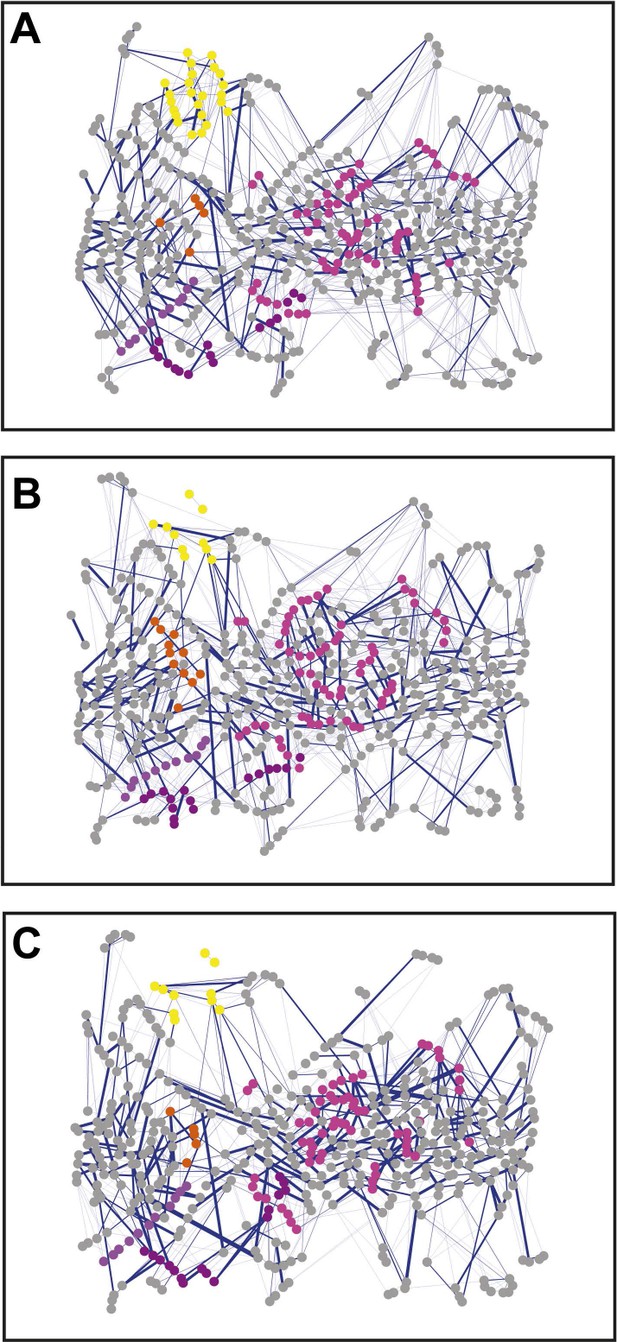
Non-covalent interaction network (non-covalent RIN) for PapC TD mutants.
Non-covalent RIN representation of the hairpin mutant (A), helix mutant (B), and helix-hairpin mutant (C) translocation domain (TD) of PapC visualized as in Figure 2.
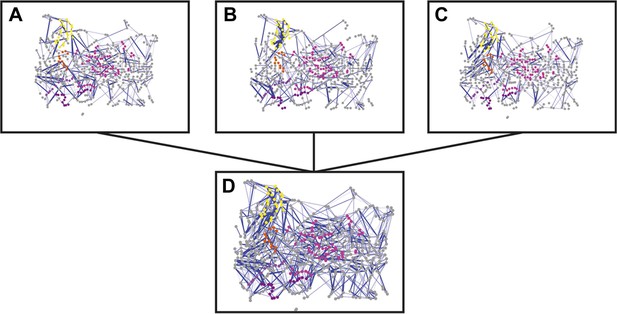
Combined RIN of the difference in non-covalent interaction score (ΔNCI score).
RINs showing the difference in non-covalent interaction score (ΔNCI score) between the native TD system and the hairpin mutant (A), helix mutant (B), and helix-hairpin mutant (C). The combined RIN (D) was created by merging the three RINs. The nodes (representing residues) are coloured as in Figure 2A. Edges (connecting two residues) are shown in blue, with edge width proportional to its corresponding ΔNCI score (from lower to higher values).
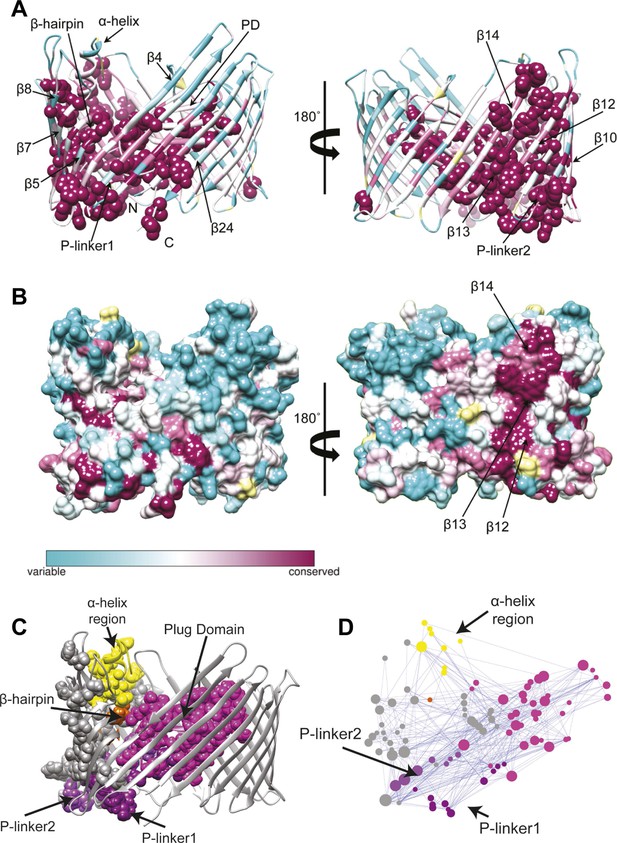
Evolutionary analysis of PapC TD.
(A–B) Sequence conservation calculated with Consurf (Ashkenazy et al., 2010) and mapped onto the initial model of the native PapC TD (sim1, t = 0). Amino acid conservation scores are classified into nine levels. The colour scale for residue conservation goes from cyan (non-conserved: grade 1) to maroon (highly conserved: grade 9), unreliable positions are coloured light yellow. (A) Ribbon representation of the model with the highly conserved residues (grade9) shown as spheres and key elements labelled. (B) Molecular surface of the model with β12–β14 labelled. (C–D) Sequence co-evolution calculated with PyCogent (Knight et al., 2007; Caporaso et al., 2008). (C) The co-evolving residues are mapped onto the initial model of the native PapC TD (sim1, t = 0). (D) The co-evolution network as visualized with Cytoscape 2.8.2 Cytoscape 2.8.2 (Smoot et al., 2011) based on RINalyzer plug-in analysis (Doncheva et al., 2011). Edges (connecting two co-evolved residues) are shown in blue, and nodes (representing coevolved residues) are coloured by structural element. The PD, P-linker1, P-linker2, β-hairpin, and α-helix are indicated schematically and coloured as in Figure 2. The node size is proportional to its degree of connectivity.
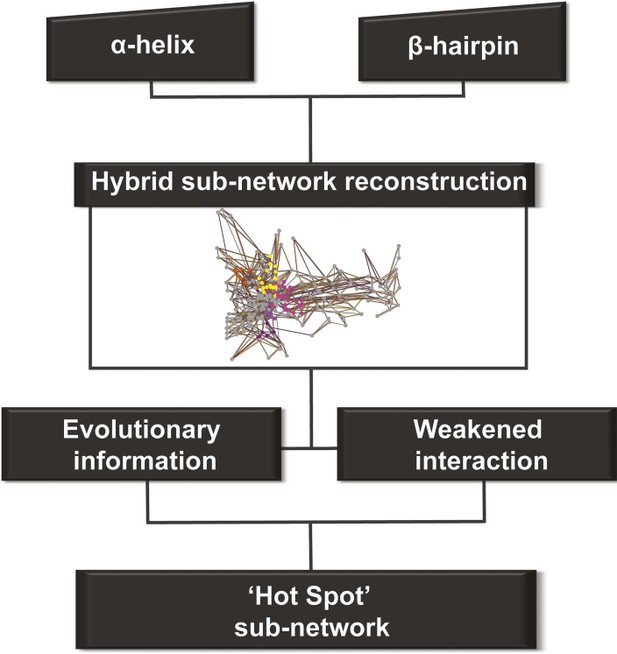
Detection of allosteric hot spots.
A flowchart representing the multistep procedure used to identify allosteric hot spots. First, a sub-network of the protein hybrid RIN was generated starting from the α-helix and β-hairpin. Then, filters based on the evolutionary information and on the interactions analysis were applied (see Figure 4—figure supplement 1) resulting in a sub-network of ‘hot spot’ residues.
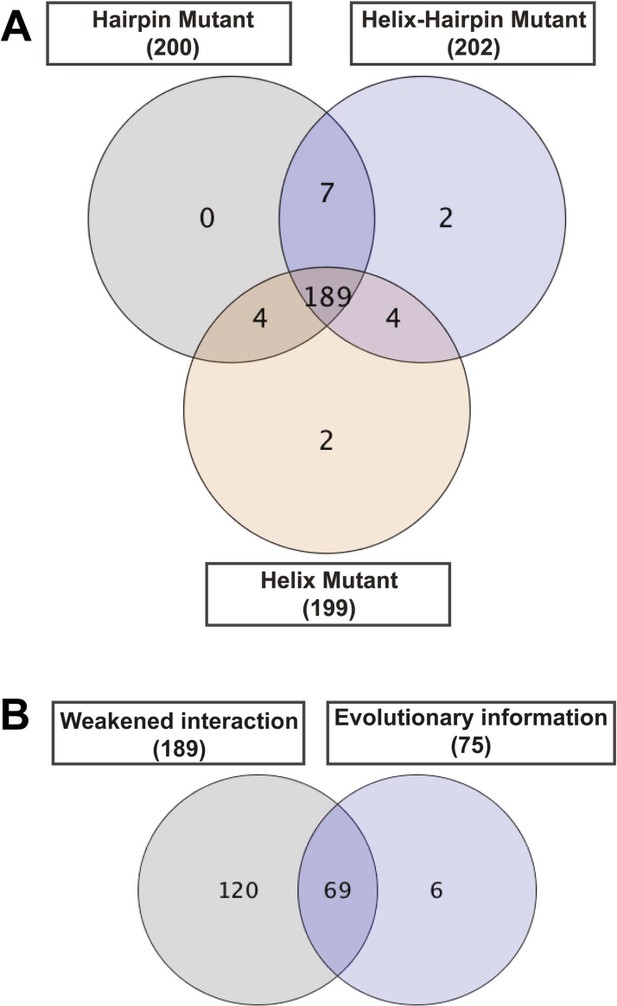
Contribution of each filter in the detection of allosteric hot spots.
Venn diagrams illustrating the contribution of each filter in the node set detection. (A) The dynamic filter resulting from the intersection of the ΔNCI score between the native TD and each of the mutants. (B) The relative combination of the dynamic filter set and the set from the evolutionary filter to the final hot spots sub-network.
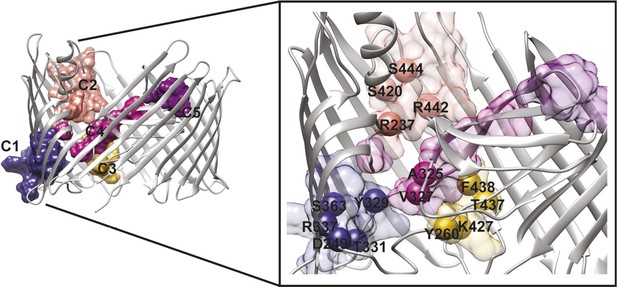
PapC TD communities.
The communities of the hot spot sub-network are shown as surface by colours and indicated schematically (C1 to C5). The inset shows a close up of the identified core residues located in β7, β8, the P-linkers, the β-hairpin, the conserved region at the base of the α-helix, in the junction between β12–β13_loop. The core residues are labelled in bold and numbered according to the X-ray structure of the apo PapC TD (PDB id: 2vqi).
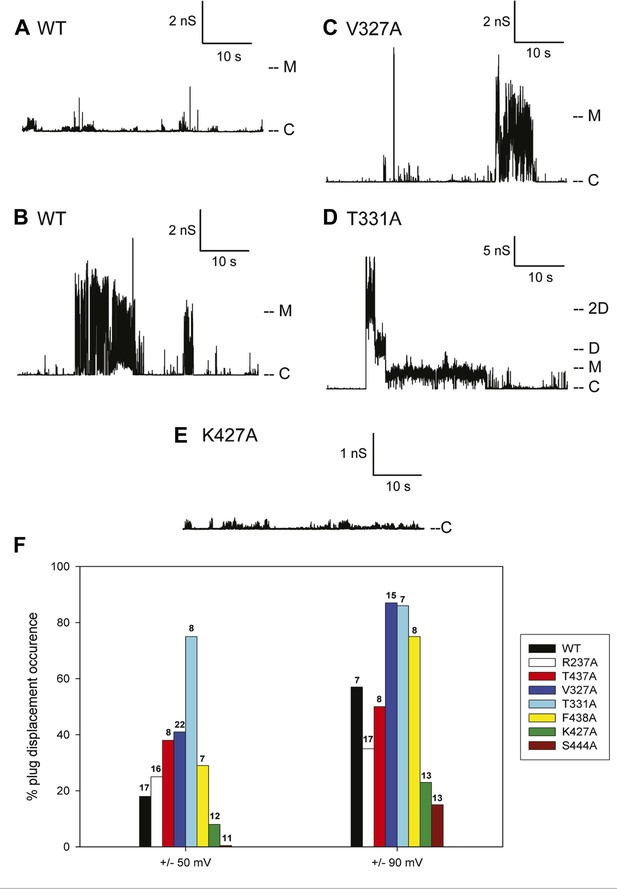
Kinetic signatures of channel activity in wildtype and mutant PapC ushers and frequency of PD displacement.
Fifty-second segments of recordings obtained in planar lipid bilayers were selected to illustrate the behaviour of the different proteins. (A) Recording from the wild-type PapC usher showing the characteristic ‘transient-mixed’ behaviour. (B) Recording from the wild-type PapC usher showing an example of spontaneous large openings due to plug displacement. Note the large amount of current fluctuations during the openings, and the ‘transient-mixed’ behaviour in between such events. Examples of similar large openings (C and D) are shown for the V327A and T331A mutants, respectively. (E) A recording from the K427A shows that the channel barely displays any activity at this voltage. The voltage was +90 mV for all panels. The current level for the closed channels is marked as ‘C’, and openings are seen as upward deflections of the traces; current levels corresponding to fully open monomeric or dimeric forms are denoted by ‘M’ and ‘D’, respectively. Note that the traces are plotted as conductance, rather than current, vs time and the scale bars are given in nS. (F) The percent of sweeps displaying ‘large-open behaviour’ (LO) indicative of PD displacement is shown for WT and each mutant at the indicated voltages. The number of individual bilayers investigated in each case is given above the bars.
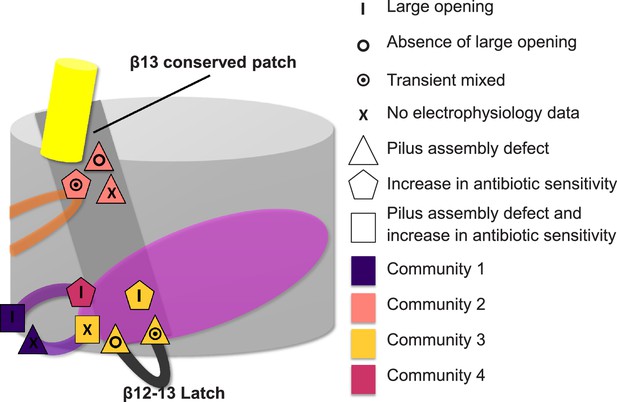
Residues involved in the allosteric signalling to control PapC gating.
A schematic model summarizing the location of the detected hot spots involved in the gating mechanism. The β12–β13_loop (the ‘latch’) and the β13 conserved patch are coloured in dark grey and light grey respectively. The PD, P-linker1, P-linker2, β-hairpin, and α-helix are coloured as in Figure 2. Hot spot residues are colour-coded based on their communities (C1–C4, as in Figure 5). The different symbols indicate the mutant's electrophysiological behaviour (‘X’ where no data were available). Mutants that show a pilus assembly defect or an increased antibiotic sensitivity, or both, are represented by a triangle, pentagon and rectangle, respectively.
Tables
Summary of the simulations.
| Simulation | Model systems | Length (ns) |
|---|---|---|
| Sim1 | Native PapC TD | 72 |
| Sim2 | Hairpin mutant | 70 |
| Sim3 | Helix mutant | 70 |
| Sim4 | Helix-hairpin mutant | 70 |
-
Descriptions of the items are: Simulation, the name of the simulation; Model systems, PapC TD model systems simulated; and Length, the length of the simulation.
Summary of the residue–residue interaction networks (RINs) parameter.
| RIN | Full RIN | C | Cr | C/Cr | L | Lr | L/Lr |
|---|---|---|---|---|---|---|---|
| Native PapC TD | 1350 (492) | 0.384 | 0.012 | 32.00 | 6.67 | 3.78 | 1.76 |
| Hairpin mutant | 1196 (485) | 0.368 | 0.011 | 33.45 | 7.20 | 3.90 | 1.84 |
| Helix mutant | 1225 (476) | 0.362 | 0.011 | 32.90 | 6.67 | 3.90 | 1.71 |
| Helix-hairpin mutant | 854 (466) | 0.262 | 0.008 | 32.75 | 8.10 | 4.70 | 1.72 |
-
Descriptions of the items are: RIN, residue–residue interaction networks of the different model systems; Full RIN, number of edges in the RIN, in parenthesis the number of node; C, average clustering coefficient; Cr, average clustering coefficient for the random networks with the same size; C/Cr, average clustering coefficient ratio (as used in Atilgan et al., 2004); L, average shortest path length; Lr, average shortest path length for the random networks with the same size; L/Lr, average shortest path length ratio (as used in Atilgan et al., 2004).
Communities in the hot spot sub-network.
| Community | Residues |
|---|---|
| C1 | E247, D249, Y329, L330, T331, G334, Q335, R337, K339, E361, S363, W364, G365, L366, S371, L372 |
| C2 | R237, D402, S420, Y441, R442, F443, S444, K468, E469, M470, E475, W496 |
| C3 | Y260, Y425, S426, K427, T437, F438, A439 |
| C4 | S233, R303, G304, L306, V308, F320, T324, A325, V327 |
| C5 | E269, E312, N314, G315, R316, K318 |
-
Descriptions of the items are: Community, the name of the community; Residues, residues that are part of the community.
Analysis of PapC substitution mutants.
| PapC | Community | HA titer | Antibiotic sensitivity | ||
|---|---|---|---|---|---|
| SDS | Erythromycin | Vancomycin | |||
| WT | 64 | 15 | 6 | 6 | |
| D249A | C1 | 0 | 15 | 6 | 6 |
| Y329A | C1 | 32 | 15 | 6 | 6 |
| T331A | C1 | 24 | 15 | 15 | 10 |
| R337A | C1 | 64 | 15 | 6 | 6 |
| S363A | C1 | 64 | 15 | 6 | 6 |
| R237A | C2 | 64 | 16 | 6 | 15 |
| S420A | C2 | 32 | 15 | 6 | 6 |
| R442A | C2 | 0 | 15 | 6 | 6 |
| S444A | C2 | 24 | 14 | 6 | 6 |
| Y260A | C3 | 0 | 15 | 14 | 6 |
| K427A | C3 | 0 | 14 | 6 | 6 |
| T437A | C3 | 24 | 14 | 6 | 6 |
| F438A | C3 | 32 | 14 | 12 | 6 |
| V327A | C4 | 64 | 20 | 14 | 16 |
-
Descriptions of the items are: PapC, the PapC construct tested; Community, the name of the community to which the mutated residue belongs; HA (hemagglutination assay) titer, the maximum fold dilution of bacteria able to agglutinate human red blood cells; Antibiotic sensitivity, the diameter of zone of inhibition (mm) around filter disc impregnated with SDS (750 µg), erythromycin (15 µg), or vancomycin (20 µg). The antibiotic sensitivity measurement includes the filter disc (6 mm diameter).
Additional files
-
Supplementary file 1
Primers used in this study to generate PapC substitution mutations.
- https://doi.org/10.7554/eLife.03532.018





