A genetically attenuated malaria vaccine candidate based on P. falciparum b9/slarp gene-deficient sporozoites
Figures
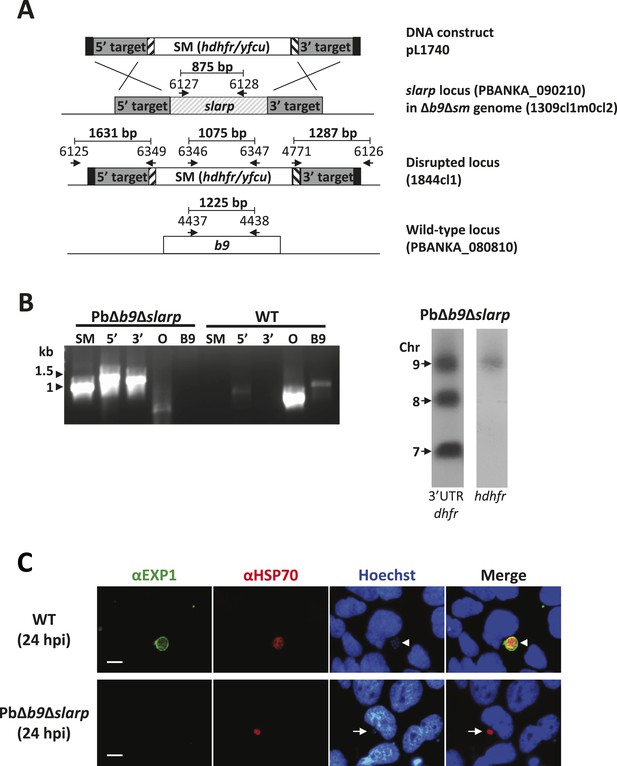
Generation and genotype analyses of P. berghei mutant PbΔb9Δslarp.
(A) Generation of mutant PbΔb9Δslarp. For PbΔb9Δslarp, the DNA-construct pL1740 was generated containing the positive/negative selectable marker cassette hdhfr/yfcy. This construct was subsequently used to generate the mutant PbΔb9Δslarp in the PbΔb9Δsm mutant. See Supplementary file 2A for the sequence of the primers. (B) Diagnostic PCR and Southern analysis of Pulse Field Gel (PFG)-separated chromosomes of mutant PbΔb9Δslarp confirming correct disruption of the slarp and the b9 locus. See Supplementary file 2A for the sequence of the primers used for the selectable marker gene (SM); 5′-integration event (5′); 3′-integration event (3′); and the slarp and the b9 ORF. For Southern analysis, PFG-separated chromosomes were hybridized using a 3′UTR pbdhfr probe that recognizes the construct integrated into P. berghei slarp locus on chromosome 9, the endogenous locus of dhfr/ts on chromosome 7, and a 3′UTR pbdhfr probe that recognizes the construct integrated into P. berghei b9 locus on chromosome 8. In addition, the chromosomes were hybridized with the hdhfr probe recognizing the integrated construct into the slarp locus on chromosome 9. (C) Development of liver-stages in cultured hepatocytes as visualized by staining with antibodies recognizing the parasitophorous vacuole membrane (anti-EXP1; green) and the parasite cytoplasm (anti-HSP70; red). Nuclei are stained with Hoechst-33342. Hpi: hours post-infection. Scale bar represents 10 µm.
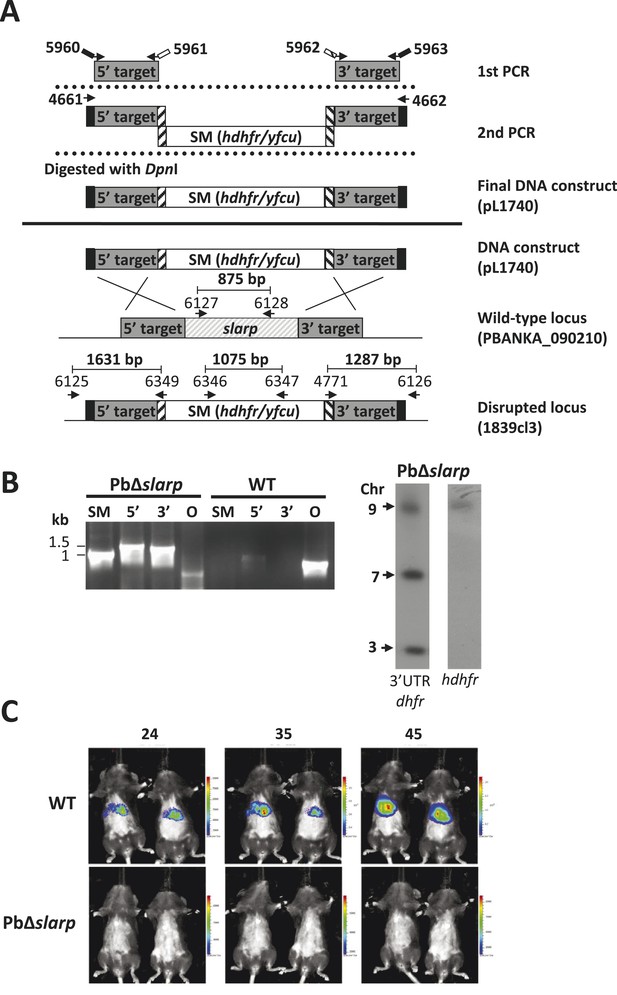
Generation and genotype analyses of P. berghei mutant PbΔslarp-a.
(A) Generation of mutant PbΔslarp-a. For PbΔslarp-a, the DNA-construct pL1740 was generated containing the positive/negative selectable marker cassette hdhfr/yfcy. This construct was subsequently used to generate the mutant PbΔslarp-a (1839cl3) in the PbGFP-Luccon reference line. See Supplementary file 2A for the sequence of the primers. (B) Diagnostic PCR and Southern analysis of Pulse Field Gel (PFG)-separated chromosomes of mutant Δslarp-a confirming correct disruption of the slarp-locus. See Supplementary file 2A for the sequence of the primers used for the selectable marker gene (SM); 5′-integration event (5′); 3′-integration event (3′); and the slarp ORF. Mutant PbΔslarp-a has been generated in the reference P. berghei ANKA line PbGFP-Luccon which has a gfp-luciferase gene integrated into the silent 230p locus (PBANKA_030600) on chromosome 3. For Southern analysis, PFG-separated chromosomes were hybridized using a 3′UTR pbdhfr probe that recognizes the construct integrated into P. berghei slarp locus on chromosome 9, the endogenous locus of dhfr/ts on chromosome 7, and the gfp-luciferase gene integrated into chromosome 3. In addition, the chromosomes were hybridized with the hdhfr probe recognizing the integrated construct into the slarp locus on chromosome 9. (C) Real time in vivo imaging of Δslarp luciferase-expressing liver-stage parasites in C57BL/6 mice at 24, 35, and 45 hr post-infection. C57BL/6 mice were IV injected with either 5 × 104 Pb-GFPLuccon sporozoites (n = 5), resulting in a full liver infection (upper panel: representative image of WT infected mice), or with 5 × 105 PbΔslarp-a sporozoites (n = 5) (lower panel: representative image of PbΔslarp-luc infected mice).
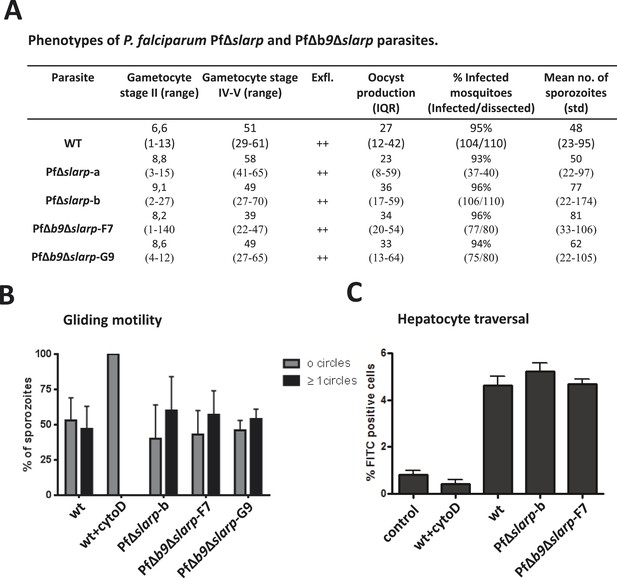
Phenotypes of P. falciparum PfΔslarp and PfΔb9Δslarp parasites.
(A) Gametocyte, oocyst, and sporozoite production. Gametocyte numbers (stage II and IV–V) per 1000 erythrocytes at day 8 and day 14 after the start of gametocyte cultures. Exflagellation (Exfl) of male gametocytes in stimulated samples from day 14 cultures (++ score = >10 exflagellation centers per microscope field at 400× magnification). Median number of oocysts at day 7, IQR is the inter quartile range and sporozoite (day 21) production (×1000) in A. stephensi mosquitoes. (B) Gliding motility of P. falciparum WT (cytochalasin D treated and untreated), PfΔslarp-b, PfΔb9Δslarp-F7, and PfΔb9Δslarp-G9 parasites. Gliding motility was quantified by determining the mean percentage ± standard deviation of parasites that exhibited gliding motility by producing characteristic CSP trails (≥1 circles) or parasites that did not produce CSP trails (0 circles). (C) Cell traversal ability of P. falciparum NF54, PfΔslarp-b and PfΔb9Δslarp-F7 sporozoites as determined by FACS counting of Dextran positive Huh7 cells. Shown is the mean percentage ±standard deviation of FITC positive cells. Dextran control (control): hepatocytes cultured in the presence of Dextran but without the addition of sporozoites.
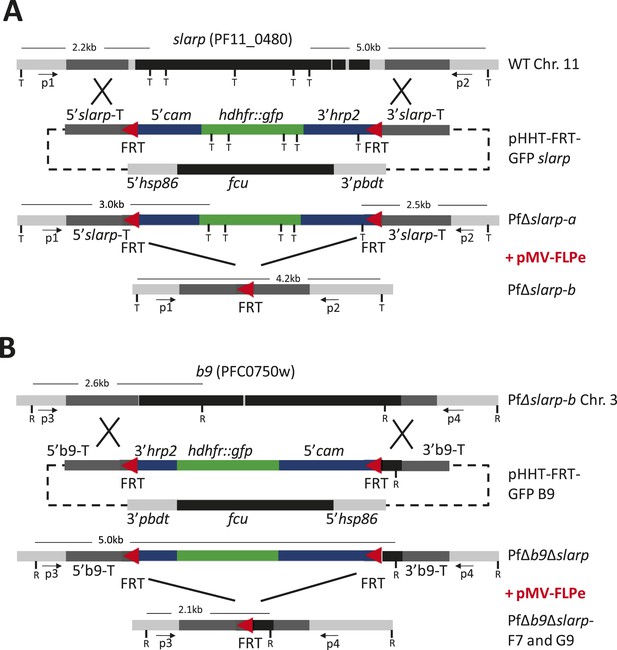
Consecutive gene deletion of slarp and b9 in P. falciparum.
Schematic representation of the genomic loci of (A) slarp (PF11_0480; PF3D7_1147000) on chromosome 11 (Chr. 11) and (B) b9 (PFC_0750w; PF3D7_0317100) on chromosome 3 (Chr. 3) of wild-type (wt; NF54wcb), PfΔslarp and PfΔb9Δslarp gene deletion mutants before (PfΔslarp a and PfΔb9Δslarp) and after the FLPe mediated removal of the hdhfr::gfp resistance marker (PfΔslarp b and PfΔb9Δslarp clones F7/G9), respectively. The constructs for the targeted deletion of slarp (pHHT-FRT-GFP slarp) and b9 (pHHT-FRT-GFP-B9) contain two FRT sequences (red triangles) that are recognized by FLPe. P1, P2 and P3, P4 primer pairs for LR-PCR analysis of slarp and b9 loci respectively; T (TaqI) and R (RcaI): restriction sites used for Southern blot analysis and sizes of restriction fragment are indicated; cam: calmodulin; hrp: histidine rich protein; hsp: heatshock protein; fcu: cytosine deaminase/uracil phosphoribosyltransferase; hdhfr::gfp: human dihydrofolate reductase fusion with green fluorescent protein; pbdt: P. berghei dhfr terminator.
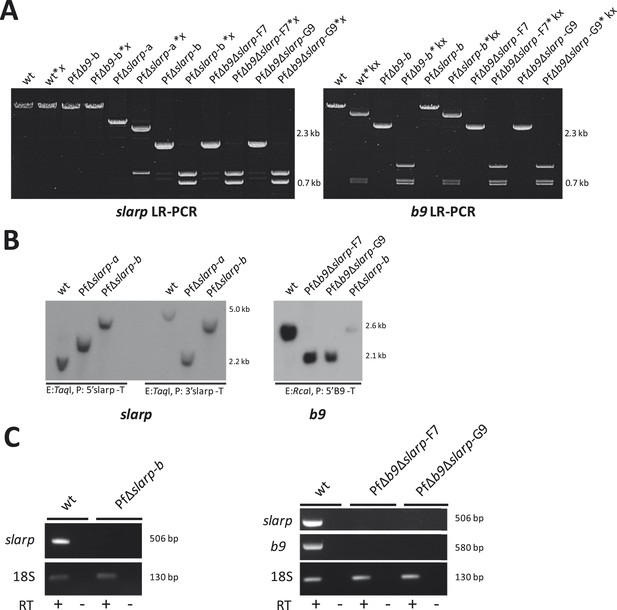
Genotype analysis of the generated PfΔslarp and PfΔb9Δslarp parasites.
(A) Long range PCR analysis of genomic DNA from WT, PfΔslarp and PfΔb9Δslarp asexual parasites confirms the slarp gene deletion and consecutive gene deletions of both slarp and b9 respectively and subsequent removal of the hdhfr::gfp resistance marker. The PCR products are generated using primers P1,P2 for slarp and P3,P4 for b9 (see A and B respectively; for primer sequences see primer table in Supplementary file 2B) and PCR products are also digested with restriction enzymes x (XmaI) and kx (KpnI/XcmI) respectively for confirmation (i.e. slarp LR-PCR product sizes: WT, 12 kb, is undigested; Δslarp-a, 5.4 kb is digested into 1.3 kb and 4.0 kb fragments, Δslarp-b, 2.4 kb is digested into 1.3 kb and 1.1 kb fragments. b9 LR-PCR product sizes: WT, 5.5 kb, is digested into 756 bp, 793 bp, and 4.0 kb fragments; Δb9-b, 2.6 kb is digested into 756 bp, 793 bp, and 1.1 kb fragments). (B) Southern analysis of restricted genomic DNA from WT, PfΔslarp-a, PfΔslarp-b, PfΔb9Δslarp-F7, and PfΔb9Δslarp-G9 asexual parasites. DNA was digested with restriction enzyme (E: TaqI) and probed with the 5′ slarp targeting region (P: 5′ slarp-T; see A) on the left side of the slarp Southern or probed with the 3′slarp targeting region (P: 3′ slarp-T; see A) on the right side of the slarp panel. For analysis of the b9, integration DNA was digested with restriction enzymes (E: RcaI) and probed with the 5′ b9 targeting region (P: 5′ b9-T; see A) on the right panel. The expected fragment sizes are indicated in panel (A). (C) RT-PCR analysis showing the absence of b9 and slarp transcripts in P. falciparum PfΔslarp-a, PfΔslarp-b, PfΔb9Δslarp-F7, and PfΔb9Δslarp-G9 mutant sporozoites. PCR amplification using purified sporozoite RNA was performed either in the presence or absence of reverse transcriptase (RT+ or RT−, respectively) and generated the expected 506 bp and 580 bp fragments for slarp and b9 respectively, the positive control was performed by PCR of 18S rRNA using primers 18Sf/18Sr (for primer sequences see Supplementary file 2B) and generated the expected 130 bp fragment.
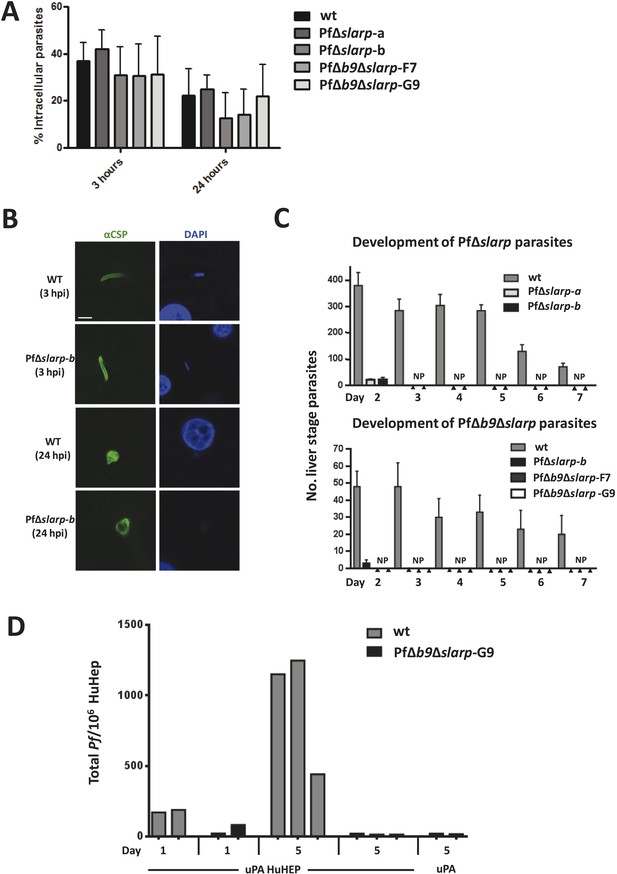
Development of P. falciparum PfΔslarp and PfΔb9Δslarp parasites in human primary hepatocytes.
(A) In vitro invasion of P. falciparum wt, PfΔslarp-a, PfΔslarp-b, PfΔb9Δslarp-F7, and PfΔb9Δslarp-G9 sporozoites in primary human hepatocytes. Invasion is represented as the mean ratio ± standard deviation of extra- and intra-cellular sporozoites by double staining at 3 and 24 hr post-infection, determined after three wash steps to remove sporozoites in suspension. (B) Immunofluorescence assay of PfΔslarp-b parasites in human primary hepatocytes at 3 and 24 hr post-infection. Parasites are visualized by staining with anti-PfCSP antibodies (green; Alexa-488) and parasite, and hepatocyte nuclei are stained with DAPI (blue). Images were photographed on an Olympus FV1000 confocal microscope. Scale bar represents 5 µm. (C) Development of P. falciparum wt, PfΔslarp-a, PfΔslarp-b (top panel), PfΔb9Δslarp-F7, and PfΔb9Δslarp-G9 (bottom panel) liver-stages in primary human hepatocytes following inoculation with 40,000 sporozoites. From day 2 to 7, the mean number ± standard deviation of parasites per 96-well was determined by counting parasites stained with anti-P. falciparum HSP70 antibodies. The bottom panel represents experiments performed in primary human hepatocytes from 2 different donors. No parasites present (NP). (D) Development of liver-stages of PfΔb9Δslarp GAP in chimeric mice engrafted with human hepatocytes. Mice were infected with 106 wt or PfΔb9Δslarp-G9 sporozoites by intravenous inoculation. At 24 hr or at 5 days after sporozoite infection, livers were collected from the mice and the presence of parasites determined by qPCR of the parasite-specific 18S DNA. uPA HuHEP; chimeric homozygous uPA+/+-SCID mice engrafted with human hepatocytes. As controls, uPA mice; heterozygous uPA+/−-SCID mice not engrafted with human hepatocytes were used.
Tables
Protection of mice after immunization with P. berghei PbΔb9 or PbΔb9Δslarp sporozoites
| Mouse strain | Pb mutant | Day of challenge* | Immunization regimes no. protected/no challenged | ||
|---|---|---|---|---|---|
| BALB/c | 10k† | 5k | 1k | ||
| PbΔb9 | 10 | 10/10‡ | 18/20 | 8/10 | |
| PbΔb9Δslarp | 10 | 20/20 | 10/10 | 20/20 | |
| C57Bl6 | 50/20/20k§ | 10/10/10k | 1/1/1k | ||
| PbΔb9 | 10 | 4/4 | nd | nd | |
| 90 | 5/5 | ||||
| 180 | 9/9# | ||||
| 365 | 5/11 | ||||
| PbΔb9Δslarp | 10 | Nd | 10/10 | 6/10 | |
| 180 | 6/6 | nd | nd | ||
-
*
Number of days post last immunization; 104 wild-type sporozoites were injected by IV route.
-
†
Immunization dose: number of sporozoites x1000.
-
‡
Protected/total # of immunized mice (%); protection was 0/15 in naive control BALB/c and 0/10 in C57BL/6 mice.
-
§
Immunization dose with 7 day intervals between immunizations.
-
#
Immunization dose 50/10/20k with 7 day intervals between immunizations. nd = not done.
Breakthrough blood-stage infections after intravenous injection of PbΔslarp and PbΔb9Δslarp sporozoites
| Mouse strain | Mutant | Infection* Spz x 103 | Breakthrough blood infection/total # mice | Pre-patent period‡ (days) |
|---|---|---|---|---|
| BALB/c | WT† | 10 | 5/5 | 4–5 |
| PbΔslarp | 50 | 0/5 | ||
| PbΔslarp | 25 | 0/10 | ||
| PbΔb9Δslarp | 25 | 0/10 | ||
| C57BL/6 | WT† | 10 | 5/5 | 4–5 |
| PbΔslarp | 500 | 0/5 | ||
| PbΔslarp | 200 | 0/10 | ||
| PbΔb9Δslarp | 200 | 0/10 | ||
| PbΔb9Δslarp | 150 | 0/5 |
-
*
Inoculation dose of sporozoites administered IV.
-
†
P. berghei ANKA strain: line cl15cy1.
-
‡
Day with parasitemia of 0.5–2%.
Additional files
-
Supplementary file 1
Oocyst and sporozoite production and sporozoite characteristics (motility, traversal, hepatocyte invasion) of the P. berghei mutants PbΔslarp and PbΔb9Δslarp.
- https://doi.org/10.7554/eLife.03582.011
-
Supplementary file 2
Primer sequences.
- https://doi.org/10.7554/eLife.03582.012





