The neural crest is a source of mesenchymal stem cells with specialized hematopoietic stem cell niche function
Figures
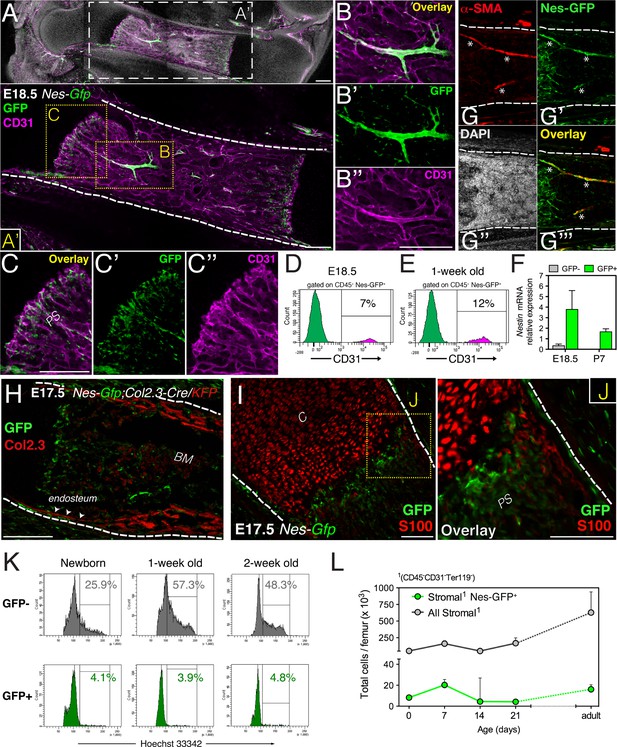
Fetal bone marrow nestin+ cells proliferate slowly and are distinct from osteochondral cells.
(A–C) Nes-GFP+ cells in fetal bones undergoing endochondral ossification. Whole-mount confocal projection of E18.5 Nes-Gfp femoral bone marrow stained with CD31 (magenta) to mark endothelium. Note the perivascular distribution of GFP+ cells (green) in arterioles (B–B′′) and small vessels invading the primary spongiosa (C–C′′). (D–E) Nes-Gfp transgene is expressed by a subset of bone marrow endothelial cells. Flow cytometry histograms show the frequency of CD45− Nes-GFP+ cells expressing CD31. (F) Endogenous Nestin mRNA expression measured by qPCR in stromal populations isolated from Nes-Gfp mice at the indicated stages (mean ± SD, n = 3–5). (G) Nes-Gfp bone marrow section stained with smooth muscle actin antibodies (αSma, red; asterisks) to reveal arterioles. (H) Limb section from an E17.5 Nes-Gfp;Col2.3-Cre;KFP embryo showing Nes-GFP+ (green) and osteoblasts identified with antibodies to Katushka (KFP) protein (red), driven by the 2.3-kb proximal fragment of the α1(I)-collagen promoter. Arrowheads, endosteal surface. (I) Metaphysis of E17.5 Nes-Gfp embryo showing S100+ chondrocytes (red). (J) Magnified view of boxed area in (I). (K) Representative cell cycle profiles of bone marrow stromal Nes-GFP+/- cells at early postnatal stages. Frequencies of cells in G2/S-M (%) are indicated. (L) Number of stromal Nes-GFP+/− cells in postnatal bone marrow (mean ± SEM, n = 3–4). Scale bars: 200 μm (A, A′, B′′, C, H), 100 μm (G, I and J); (A′, G–J) dashed line indicates bone contour. BM, bone marrow; C, cartilage; PS, primary spongiosa.
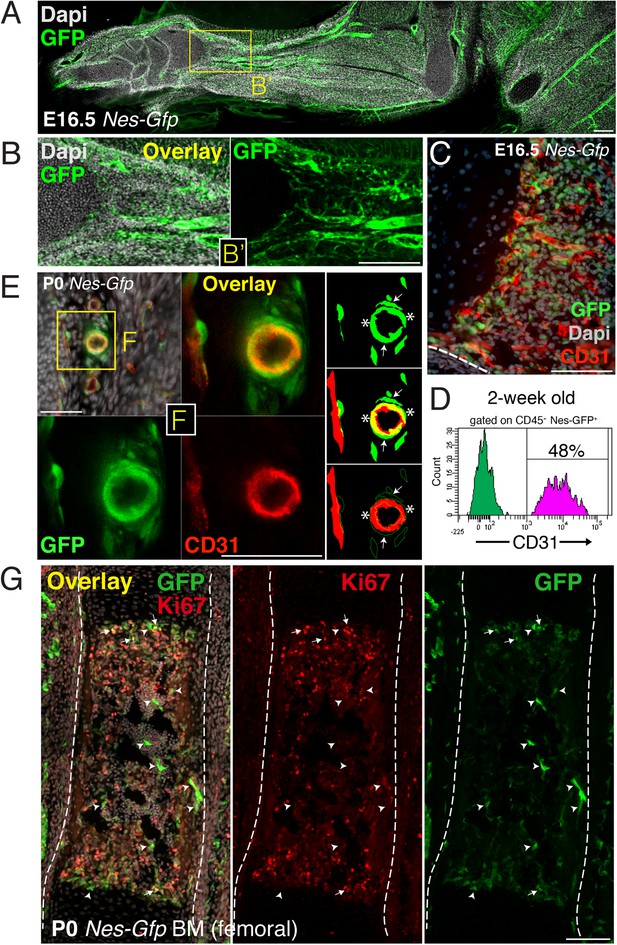
Perivascular and endothelial Nes-GFP+ cells invade the incipient bone marrow associated with blood vessels.
(A) Longitudinal section of E16.5 Nes-Gfp forelimb. A profuse perivascular network of GFP+ cells can be observed, mostly associated with arterial blood vessels. (B) High magnification detail (inset B’) of arterioles containing Nes-GFP+ cells during invasion of the distal shaft of fetal bone. (C) Section through an E16.5 Nes-Gfp metaphyseal region undergoing vascularization, revealed by CD31 immunostaining of endothelial cells (red). (D) Representative FACS histograms of anti-CD31-stained CD45- bone marrow cells from 2-week old Nes-Gfp mice. The frequency of GFP+ putative endothelial cells is indicated. (E and F) Confocal high magnification detail is showing several layers of perivascular GFP+ cells (arrows) encircling an arteriole. Innermost endothelial cells immunostained with CD31 (red) also expressed GFP (yellow overlay, asterisks). (G) Femoral bone marrow section from newborn Nes-Gfp mouse stained with Ki67 (red) to label proliferative cells. Arrows indicate GFP+ Ki67+ cells; arrowheads depict GFP+ Ki67− cells; dashed line marks bone contour. (A-C,E,G) Nuclei were counterstained with DAPI (gray). Scale bars: 500 μm (A); 200 μm (B); 100 μm (C,G); 50 μm (E and F).
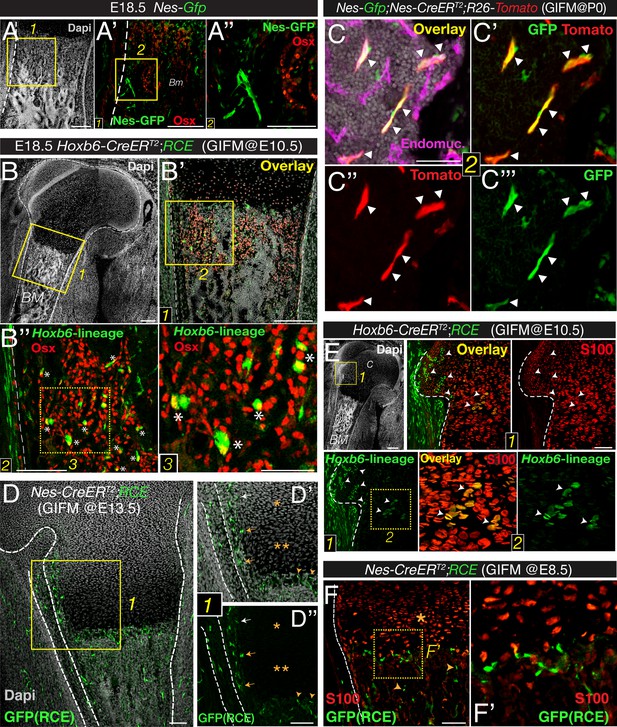
Bone marrow nestin+ cells are different from mesodermal osteo-chondroprogenitors.
(A and B) Bone marrow sections from Nes-Gfp (A–A′′) and Hoxb6-CreERT2;RCE (B–B′′) E18.5 embryos (tamoxifen-induced at E10.5) immunostained with Osterix antibodies (Osx, red) to label osteoprogenitor cells. GFP+ Osx+ mesodermal-derived osteoprogenitors are marked with asterisks (insets 2–3). (C–C′′′) Perinatal recombination in Nes-CreERT2 mice efficiently targets bone marrow stromal Nes-GFP+ cells. Bone marrow section of a P7 Nes-Gfp;Nes-CreERT2;R26-Tomato mouse that received tamoxifen at birth, showing Nes-GFP+ cells (green), Nes-derived progeny (red), and double-positive cells (arrowheads). (D–F) Fate mapping of the progeny of nestin+ cells and limb mesoderm in E18.5/19.5 femoral bone marrow from Nes-CreERT2;RCE (D–D′′, F–F′) and Hoxb6-CreER;RCE fetuses (E). (D) GFP (green) and nuclei counterstained with DAPI (gray) in bone of E18.5 fetus induced with tamoxifen at E13.5. Neither proliferating (*) nor hypertrophic (**) chondrocytes showed GFP fluorescence (inset 1). (D′–D′′) Nes-derived cells with a similar morphology and distribution to Nes-GFP+ cells were detected near the cartilage–perichondrium interface (arrows) and within the chondro–osseous junction (arrowheads). (E and F) Bone marrow sections of (E) Hoxb6-CreER;RCE and (F) Nes-CreERT2;RCE E18.5 embryos induced with tamoxifen at E10.5 and E8.5, respectively, stained with S100 antibodies to label chondrocytes (red). High magnification views of cartilage (inset 2) showing abundant double-positive chondrocytes (arrowheads). (F) Nes-traced cells (green) were not chondrocytes (red, *) but infiltrated the chondro–osseous junction and trabecular bone (arrowheads). Scale bars: 200 μm (A–A′, B–B′), 100 μm (A′′, B′′), 50 μm (B′′3, C). BM, bone marrow; C, cartilage; GIFM, genetic inducible fate mapping.
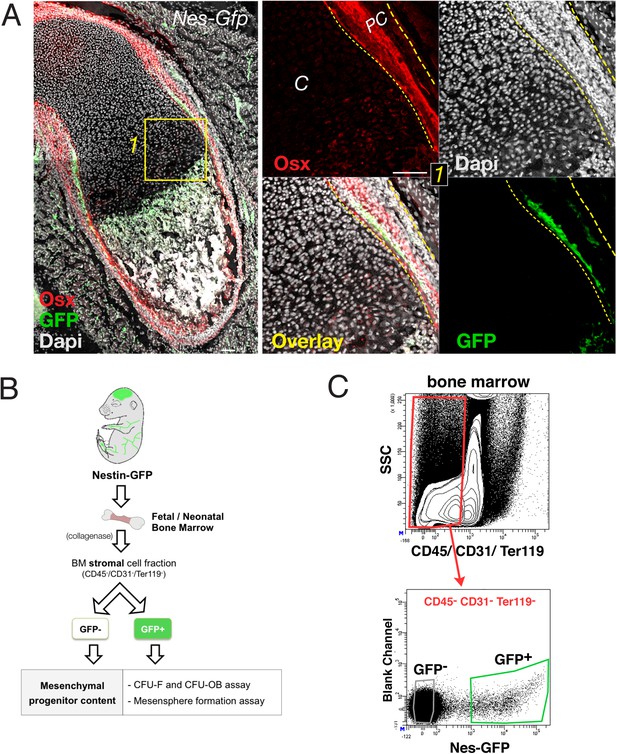
Sub-fractionation of fetal bone marrow mesenchymal progenitors.
(A) Neonatal Nes-Gfp bone marrow section immunostained with anti-osterix antibodies (red). Right panels, perichondrium detail showing inner rim of GFP+ cells (green) adjacent to distinctive perichondrial osterix+ cells. Nuclei were counterstained with DAPI (gray). Scale bar, 100 μm. PC, perichondrium. (B) Scheme showing FACS isolation of bone marrow stromal populations from Nes-Gfp mice and MSC assays. (C) Representative FACS plots and gating strategy to isolate stromal GFP+/- cells from Nes-Gfp bone marrow.
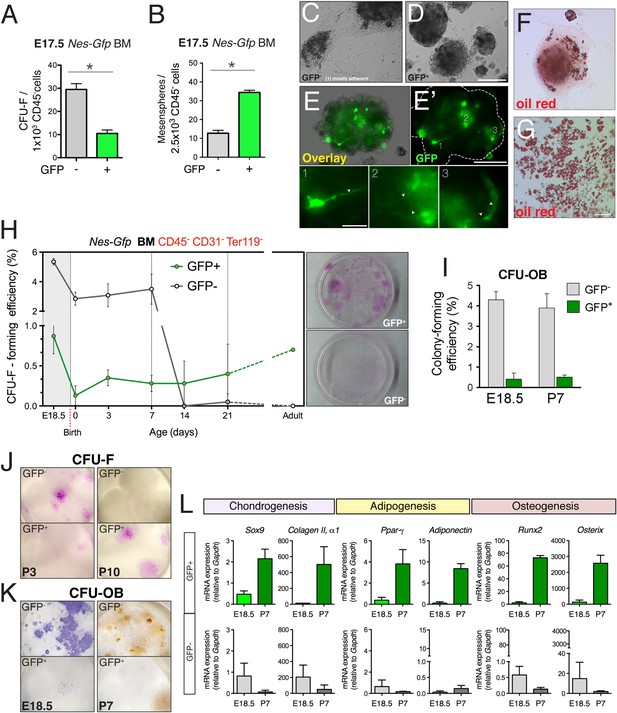
Perinatal enrichment of MSC activity in bone marrow nestin+ cells.
(A and B) Fibroblast colony-forming units (CFU-F) and mesensphere-forming activities segregate in Nes-GFP- and Nes-GFP+ fetal bone marrow cells, respectively. Frequencies of CFU-F and mesensphere-forming efficiency in E17.5 Nes-Gfp embryos. (C–E) Representative sphere cultures from both mesenchymal subpopulations sorted from bone marrow of Nes-Gfp fetuses. Note the presence of GFP+ fibroblast-like cells (E′ and insets1–3). (F–G) Adherent colonies derived from GFP- population stained with Oil Red O (red), to reveal mature adipocytes. (H) MSC activity is progressively restricted to bone marrow Nes-GFP+ cells. Frequency of CFU-Fs in cultures of stromal (CD45− CD31− Ter119−) GFP+/− cells, isolated from the bone marrow of Nes-Gfp mice of the indicated age. Right panels show representative CFU-Fs in cell populations from adult mice. (I) Frequency of osteoblastic colony-forming units (CFU-OB) in bone marrow stromal GFP+/− cells of the indicated age. (J) Representative Giemsa-stained CFU-F from 3- and 10-day old bone marrow subpopulations. (K) Stained CFU-OB from E18.5 (alkaline phosphatase staining, left panels) and 1-week old (alizarin red staining, right panels) bone marrow subpopulations. (L) qPCR analysis of mesenchymal genes in bone marrow stromal populations isolated from fetal (E18.5) or 1-week old (P7) Nes-Gfp mice, as depicted (Figure 3—figure supplement 1). (A–B, H–L) Mean ± SD, n = 3–6; *p < 0.05, unpaired two-tailed t test. Scale bars: 200 μm (D, E′, G), 100 μm (G), 50 μm (E′1–3).
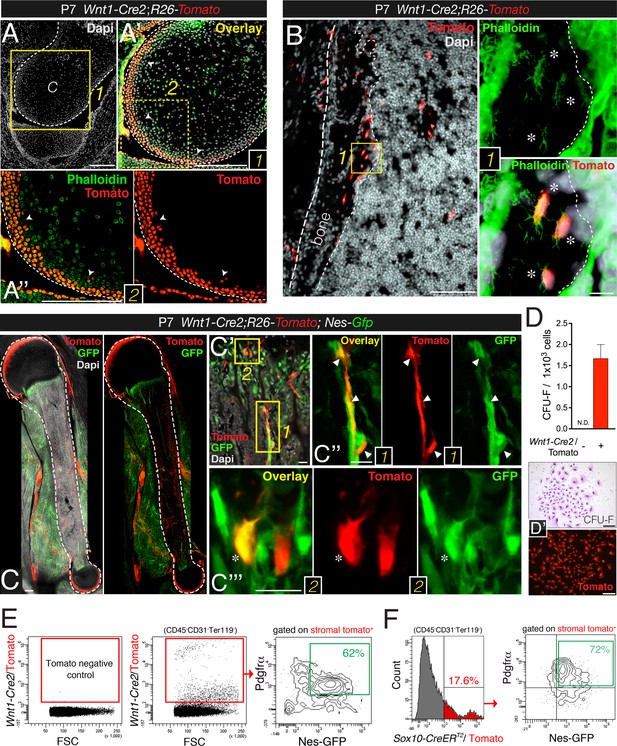
Contribution of trunk neural crest cells to mesenchymal lineages in long bones.
(A and B) Fate mapping of neural crest derivatives in femoral bone marrow of neonatal Wnt1-Cre2;R26-Tomato mice. (A) Section through femoral distal epiphysis showing cortical neural-crest-derived chondrocytes (arrowheads, red); green signal corresponds to phalloidin staining. Nuclei were counterstained with DAPI (gray). (B) Bone marrow section showing Wnt1-Cre2-derived Tomato+ (red) osteocytes. (Inset 1) Neural-crest-derived osteocytes (asterisks) in endosteal region, showing their typical morphology revealed by phalloidin staining (green). (C–E) The neural crest contributes to Pdgfrα+ BMSCs in long bones. (C–C′′) Fluorescent signals of GFP, Tomato, and DAPI in bone marrow sections from 1-week old Wnt1-Cre2;R26-Tomato;Nes-Gfp mice. (D) Frequency of fibroblastic colony-forming units (CFU-F) in CD31- CD45- Ter119- Tomato+/- bone marrow cells sorted from 1-week old Wnt1-Cre2;R26-Tomato mice (n = 3); N.D., not detectable. (D′) Examples of Giemsa staining (top panel) and Tomato fluorescence in neural crest-derived CFU-Fs. (E) Representative flow cytometry analysis of bone marrow stromal cells from 4-week old Wnt1-Cre2;R26-Tomato;Nes-Gfp mice. (F) Flow cytometry analysis of bone marrow stromal cells from Nes-Gfp;Sox10-CreERT2;R26-Tomato triple-transgenic mice stained with Pdgfrα antibody. (E, F) Frequencies of neural crest-traced BMSCs are indicated. Scale bars: 200 μm (A–A′′, C, D–D′), 100 μm (B), 20 μm (B1, C′–C′′′). Dashed line depicts the bone and cartilage contour (A–C).
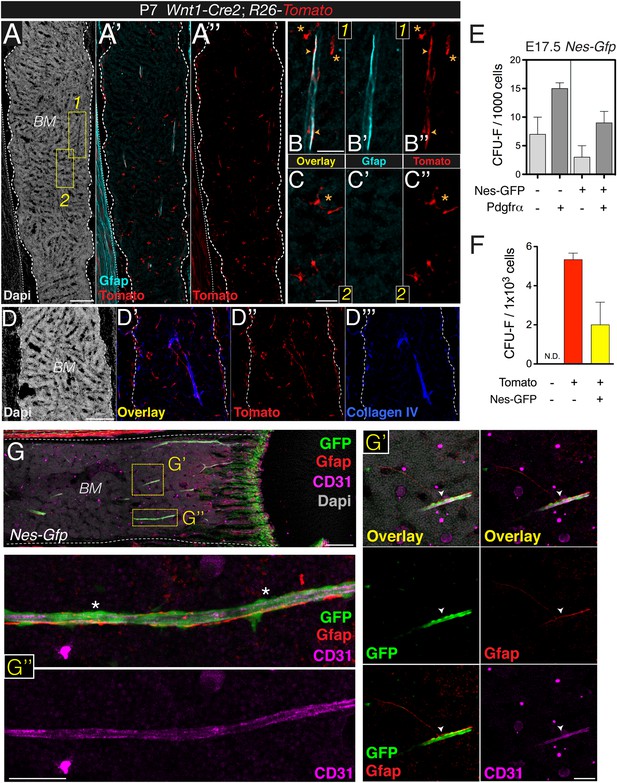
Bone marrow Nes-GFP+ cells are distinct from mature Schwann cells.
(A–C) Diaphysis section stained with antibodies against glial fibrillary acidic protein (Gfap, cyan). Neural crest-traced cells included Gfap+ Schwann cells (inset 1, arrowheads) and Gfap− putative BMSCs (inset 2, asterisks) (B-B′′, C-C′′). (D-D′′′) Staining of basal lamina with collagen IV antibodies (blue) showing close association of neural crest-derived cells (red) with blood vessels. (E) Surface expression of Pdgfrα enriches for fetal bone marrow mesenchymal progenitors. Fibroblast colony-forming units (CFU-F) in bone marrow stromal cells (CD45- CD31- Ter119-) isolated from E17.5 Nes-Gfp mice according to GFP and Pdgfrα expression. (F) Frequency of CFU-F colonies obtained from sorted populations of Wnt1-Cre2;R26-Tomato;Nes-Gfp 1-week old pups. (G) Schwann cells are closely associated with distinctive Nes-GFP+ perivascular cells (arrowheads). Immunofluorescence of P9 Nes-Gfp tibial bone marrow showing GFP+ cells (green), Gfap+ Schwann cells (red), and CD31+ endothelial cells (pink); nuclei were counterstained with DAPI (gray). (G′–G′′) Details of central diaphyseal region (insets) at high magnification. (G′) Note the long Gfap+ Schwann cells extending through the arteriole, surrounded by distinctive Nes-GFP+ cells (arrowhead). (G′′) Magnified view of a long arteriole. Asterisks indicate Gfap- perivascular Nes-GFP+ cells. Scale bars: 200 μm (A, D, G), 50 μm (B, C, G′–G′′).
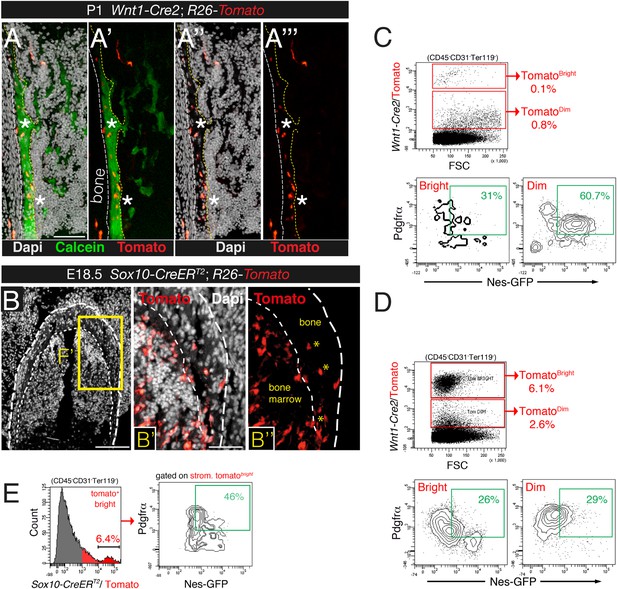
Contribution of trunk neural crest to bone marrow stromal lineages.
(A-A′′′) Bone marrow section from newborn Wnt1-Cre2;R26-Tomato pup stained with calcein to mark calcium deposition (green), showing Wnt1-Cre2-traced Tomato+ osteoblasts (red) in calcifying areas (asterisks). Scale bar = 100 μm. (B) Representative bone section of an E18.5 Sox10-CreERT2;R26-Tomato mouse showing Tomato+ osteocytes (asterisks) and bone-lining osteoblasts (red). Dashed line depicts the bone contour. Nuclei were counterstained with DAPI (gray). (C and D) Representative flow cytometry plots of bone marrow stained with Pdgfrα from neonatal (D) and 4-week old (C) Wnt1-Cre2;R26-Tomato;Nes-Gfp mice, after gating on the stromal (CD45- CD31- Ter119-) population. (E) Representative flow cytometry histogram of bone marrow cells from E18.5 Sox10-CreER;R26-Tomato;Nes-Gfp embryos, induced with tamoxifen at E9.5, after gating on the tomato-bright population. Scale bars: 200 μm (B), 100 μm (A, B′).
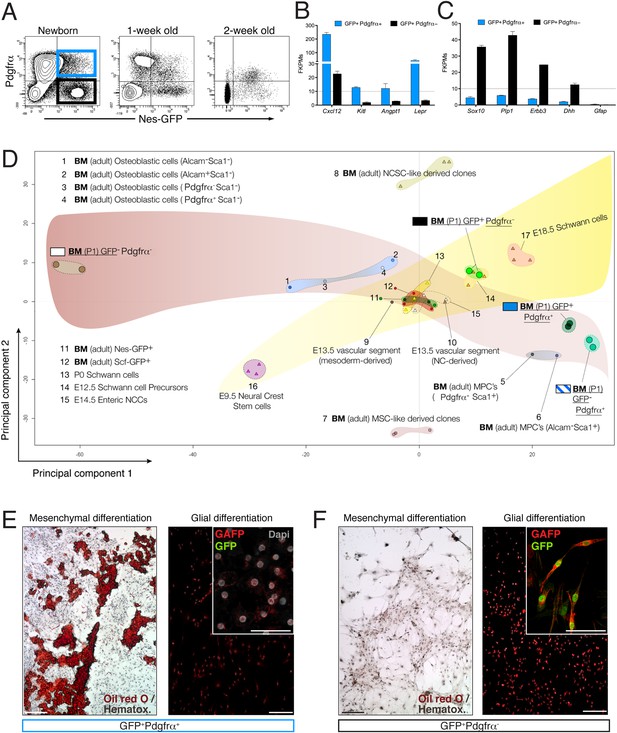
The neonatal bone marrow Nestin-GFP+ population contains Pdgfrα+ MSCs and Pdgfrα− Schwann cell precursors.
(A) Representative flow cytometry profiles showing Nes-GFP and Pdgfrα expression in postnatal BMSCs. (B and C) Relative mRNA expression levels of (B) HSC niche-related genes and (C) Schwann cell progenitor genes by GFP+ Pdgfrα+ (black) or GFP+ Pdgfrα− BMSCs. RNAseq data are expressed as fragments per kilobase of exon per million fragments mapped (FPKM; n=2 independent samples from pooled newborns). Note that the neonatal GFP+ Pdgfrα− subpopulation has a Schwann cell progenitor signature (C), whereas GFP+ Pdgfrα+ cells are enriched in HSC maintenance genes. (D) Principal component analysis comparing the transcriptome of neonatal Nes-Gfp bone marrow stromal subsets with available microarray expression data sets from neural crest-derived populations and primary adult mouse BMSCs (Table 1). (E and F) In vitro differentiation of neonatal subpopulations isolated as in (A) and cultured in mesenchymal (mesenchymal) and Schwann cell (glial) differentiation medium. Adipocytes were stained with Oil Red O (red) and counterstained with hematoxylin (left panels); Schwann cells were stained with antibodies against glial fibrillary acidic protein (Gfap, red) and overlaid with endogenous GFP fluorescence (right panels). Scale bars: 200 μm (top right insets: 50 μm).
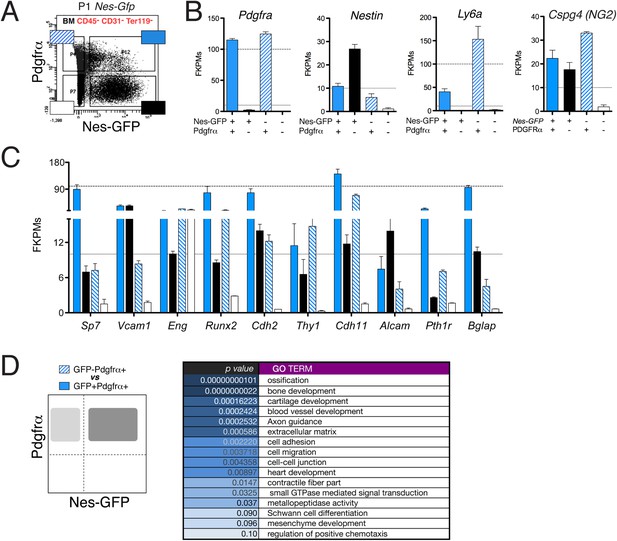
The neonatal Nes-GFP+ bone marrow population is enriched in primitive mesenchymal progenitors.
(A) Representative FACS profile of GFP and Pdgfrα expression in CD45− CD31− Ter119− BMSCs from neonatal Nes-Gfp long bones. Four different stromal subpopulations identified based on expression levels of Pdgfrα and GFP were isolated and analyzed by RNAseq. (B) Endogenous transcript expression levels of Nestin, Pdgfra, Cspg4 (NG2), and Sca1 (Ly6a) in isolated stromal populations depicted in (A), expressed in fragments per kilobase of exon per million fragments mapped (FPKM). (C) Expression profiles of characteristic mesenchymal genes in the stromal subpopulations. (D) Functional gene ontology enrichment analysis (DAVID software) of genes differentially expressed (p ≤ 0.10) between the GFP+ and GFP− subsets of P1 Pdgfrα+ bone marrow stromal cells.
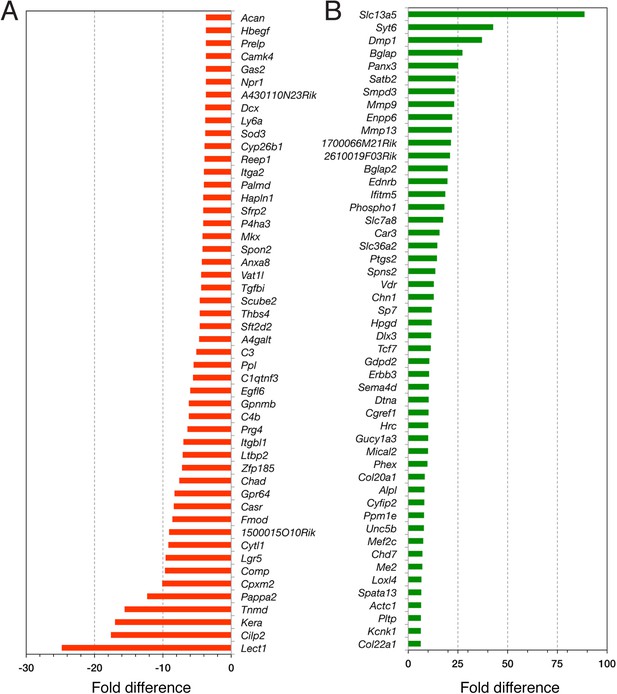
RNA-seq data analysis.
Top 50 downregulated (A) and upregulated (B) genes in Nes-GFP+ Pdgfrα+ and Nes-GFP+ Pdgfrα− cells from P1 bone marrow stroma.
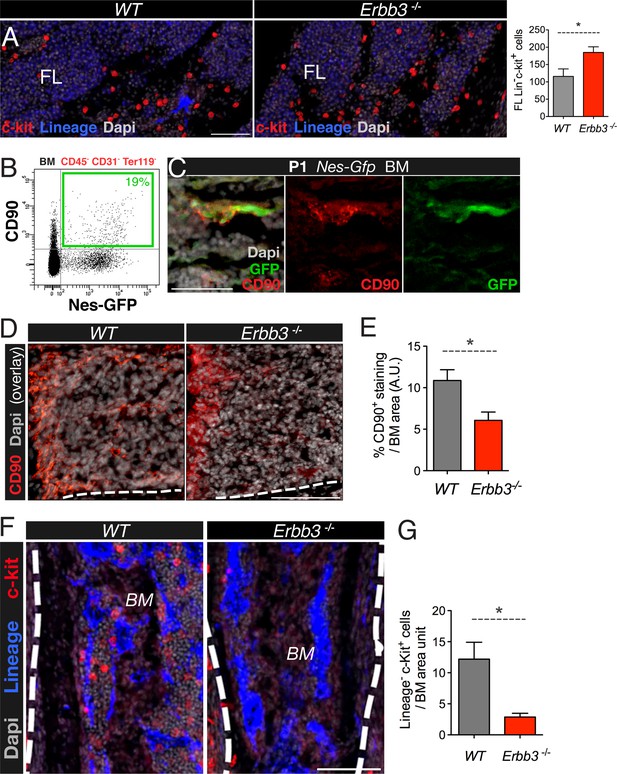
Perineural migration of neural crest-derived cells to long bones generates nestin+ MSCs with specialized HSC niche function.
(A) E17.5-E18.5 fetal liver (FL) sections from wild-type (wt) and Erbb3-null embryos stained with antibodies for mature hematopoietic lineage (blue) and c-Kit (red). Quantification of fetal liver Lin− c-Kit+ hematopoietic progenitors (per 0.41 mm2). (B) Representative FACS profile of CD45- CD31-Ter119- bone marrow cells from 2-week old Nes-Gfp mice stained with the mesenchymal marker CD90, showing the expression enrichment in Nes-GFP+ cells. (C) Neonatal bone marrow section stained with anti-CD90 (red), which labeled Nes-GFP+ (green) cells. Scale bar: 50 μm. (D) Representative bone marrow sections from wt and Erbb3-null E17.5/18.5 mice immunostained with anti-CD90 (red). (E) Quantification of CD90 immunostaining of samples in (D); n = 3. (F) Staining of bone marrow sections from wt and Erbb3-null embryos with antibodies for mature hematopoietic lineage (blue) and c-Kit (red). (G) Quantification of bone marrow Lineage− c-Kit+ hematopoietic progenitors in E17.5/18.5 wt and Erbb3-null mice (n = 3). (E, G) Mean ± SEM; *p < 0.05, unpaired two-tailed t test.
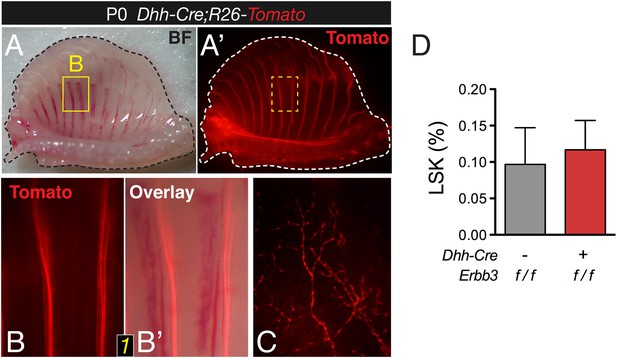
Conditional Erbb3 deletion after glial specification of Schwann cell precursors does not affect bone marrow HSCs.
(A-A′) Whole-mount view of ventral ribcage of a Dhh-Cre;R26-Tomato neonate, showing labeling of glial cells along the intercostal peripheral nerves (red). (B-B′) Surface detail of Tomato+ Schwann cells between 2 ribs. (C) High magnification of ventral skull showing an intricate network of Tomato+ Schwann cells. (D) Frequency of hematopoietic lineage− c-Kit+ Sca-1+ (LSK) cells in neonatal bone marrow of Dhh-Cre;Erbb3f/f and control littermate mice (mean ± SD, n = 4–5).
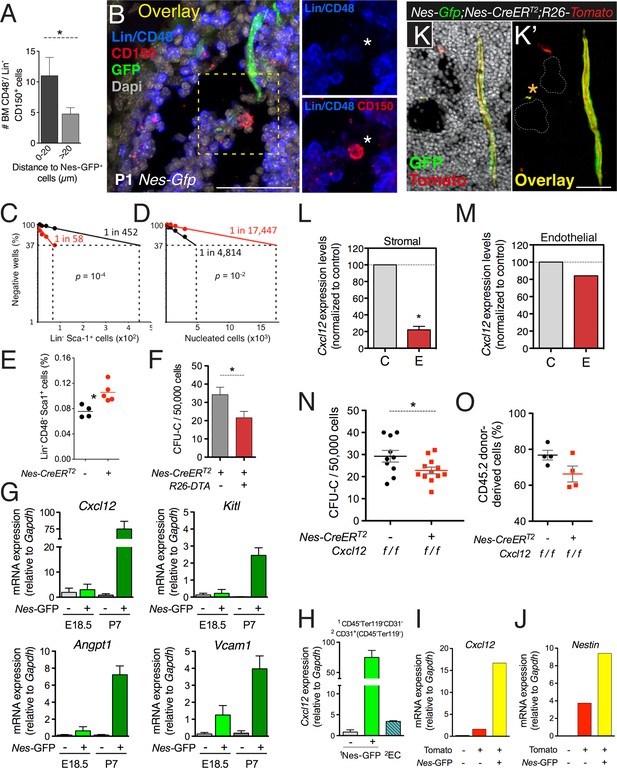
CXCL12 produced by nestin+ MSCs contributes to the establishment of the HSC niche in the bone marrow.
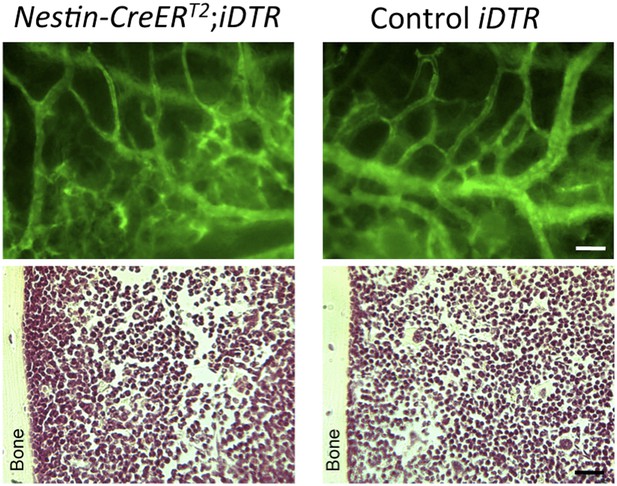
(A and B) HSCs are localized near Nes-GFP+ cells in neonatal bone marrow. Neonatal femoral sections from Nes-Gfp mice were immunostained with antibodies for mature hematopoietic lineages, CD48 (blue) and CD150 (red). (A) Quantification of the distance of Lin− CD48− CD150+ HSC-enriched cells from Nes-GFP+ cells (mean ± SEM, n = 41). (B) Representative image of a putative HSC (asterisk) near a Nes-GFP+ cell. (C–F) Depletion of nestin+ cells compromises developmental HSC migration to bone marrow. (C and D) Long-term culture-initiating cell (LTCIC) assay from nestin-depleted fetal liver and bone-marrow cells. Nes-CreERT2;iDTR (red dots) and control iDTR (black dots) mice were exposed to tamoxifen at E14.5 and diphtheria toxin at E15.5, and liver cells (C) and bone marrow cells (D) were isolated at E17.5 (n = 5-6). The percentage of culture dishes that failed to generate hematopoietic colony-forming units in culture (CFU-C) is plotted against five serial dilutions of (C) fetal liver Lin− Sca-1+ cells and (D) nucleated bone marrow cells. HSC frequencies and p values are indicated (Pearson’s chi-squared test). (E) Frequency of Lin− Sca-1+ E17.5 liver cells in mice in (C). (F) Bone marrow CFU-C content in 1-week old Nes-CreERT2;R26-DTA and control littermates treated with tamoxifen at birth (n = 3–7). (G) Expression of core HSC maintenance genes increases in perinatal Nes-GFP+ BMSCs. qPCR analysis of Cxcl12, stem cell factor/kit ligand (Kitl), angiopoietin-1 (Angpt1), and vascular cell adhesion molecule-1 (Vcam1) mRNA in CD45- CD31- Ter119- GFP+/− cells isolated from E18.5 and P7 Nes-Gfp bone marrow. (H) Relative Cxcl12 mRNA expression levels in endothelial cells and Nes-GFP+/- BMSCs isolated from 1-week old mice (qPCR; n = 2). (I and J) Relative enrichment of Cxcl12 (I) and Nestin (J) mRNA expression in populations sorted from the bone marrow of P7 Nes-Gfp;Wnt1-Cre2;R26-Tomato compound transgenic mice. (K) Representative confocal image of a bone marrow section from a 1-week old (P7) Nes-Gfp;Nes-CreERT2;R26-Tomato mouse treated with tamoxifen at birth. Both sinusoidal (asterisk) and arteriolar GFP+ cells express the Nes-CreERT2-derived Tomato (red) reporter (yellow in overlaid picture, K′). (L and M) Efficiency of perinatal Cxcl12 excision by the Nes-CreERT2 driver in CD45-Ter119−CD31- cells (L) and endothelial (M) cells isolated from P7 bone marrow; qPCR in CD45-Ter119−CD31- cells isolated from Cxcl12f/f;Nes-CreERT2 (E) and control (C) littermates treated with tamoxifen at birth (n = 2-3). (N and O) Bone marrow CFU-C (N) and long-term HSC (O) content in P7 Cxcl12f/f;Nes-CreERT2 and control littermates treated with tamoxifen at birth. (O) Lethally-irradiated mice (CD45.1) were transplanted with 1 ×106 bone marrow cells from P7 Cxcl12f/f;Nes-CreER or Cxcl12f/f mice (CD45.2), together with 1 × 106 recipient bone marrow cells (CD45.1). Peripheral donor-derived blood chimerism after 16 weeks is shown (n = 4 per group). (E, L) Each dot represents an individual mouse. (F, H–J) Mean ± SD. *p < 0.05, unpaired two-tailed t test.
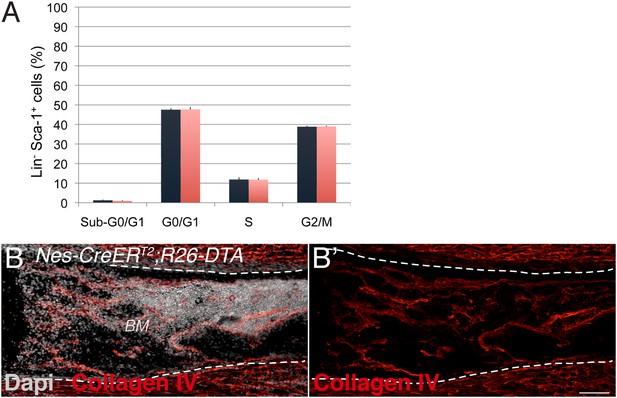
Neural crest-derived cells direct developmental HSC migration to the bone marrow.
(A) Cell cycle profile of Lin− Sca-1+ E17.5 liver cells isolated from Nes-CreERT2;iDTR embryos and control iDTR littermates 24 hr after tamoxifen administration and 48 hr after diphtheria toxin treatment (mean ± SEM, n = 6). (B) Representative femoral bone marrow section from a tamoxifen-treated Nes-CreERT2;R26-DTA mouse immunostained with collagen IV antibody (red) to reveal blood vessels (B′). Nuclei were counterstained with DAPI (gray). Scale bar, 100 μm.
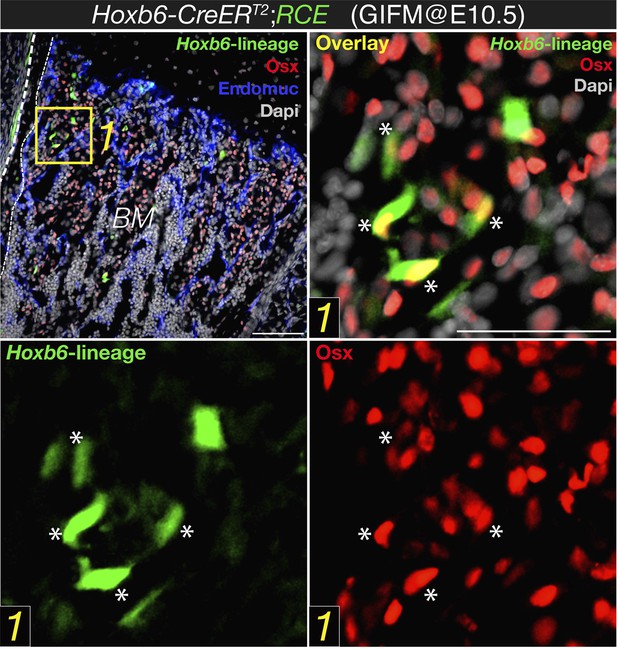
Expression of osterix by bone marrow cells of the Hoxb6-derived lineage. High magnification detail of E18.5 Hoxb6-CreERT2;RCE bone marrow (BM) stained with anti-osterix (red). Embryos were induced with tamoxifen at E10.5 to trace limb mesoderm with Hoxb6-Cre driver. Asterisks indicate nuclear osterix-positive cells also marked with GFP. Dashed line indicated the bone contour. Scale bar: 50μm.
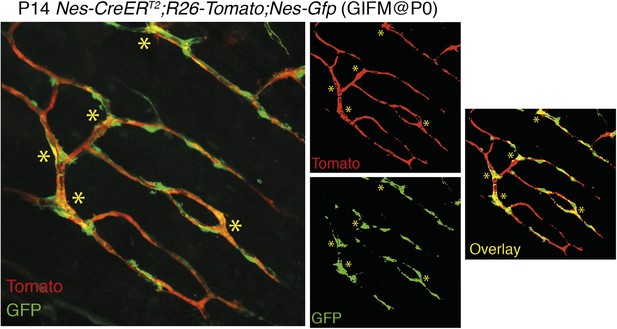
Overlap of Nestin-GFP+ cells with Nestin-CreERT2-labelled cells in the bone marrow. Representative confocal projections of skull bone marrow, showing the endogenous fluorescent signals of Tomato in red (upper right) and GFP in green (lower right); their corresponding overlay is indicated in orange/yellow. Nestin-gfp;Nestin-CreERT2;R26-Tomato triple-transgenic mice were treated with tamoxifen (4 mg, oral gavage to mother) at neonatal stage (P0) and were analyzed after 2 weeks. Asterisks indicate double-positive cells.
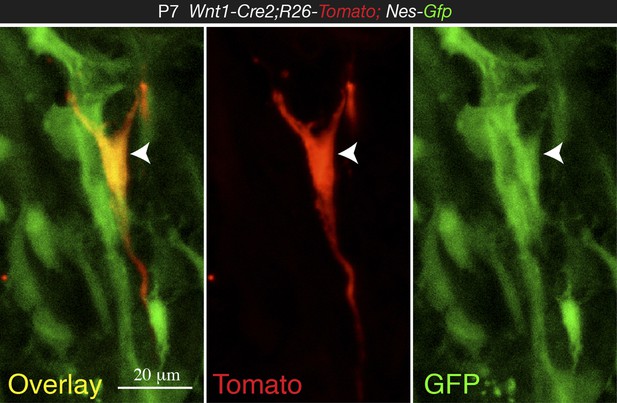
High magnification confocal image from 1-week old Wnt1-Cre2;Tomato;Nes-Gfp BM section, showing co-localization of NC-derived Tomato+ cell with Nes-GFP (arrowhead).
Tables
Description of publicly available data sets used for principal component analyses
| Plot ID | Cell Population | GEO* samples | Sample description | Ref |
|---|---|---|---|---|
| 1 | BM osteoblastic cells (Alcam- Sca1-) | GSM437794 | BM (adult) primary stromal† Alcam- Sca1- | ‡ |
| 2 | BM osteoblastic cells (Alcam+ Sca1-) | GSM437795 | BM (adult) primary stromal† Alcam+ Sca1- | |
| 3 | BM osteoblastic cells (Pdgfrα- Sca1-) | GSM437797 | BM (adult) primary stromal† Pdgfrα- Sca1- | |
| 4 | BM osteoblastic cells ( Pdgfrα+ Sca1-) | GSM437798 | BM (adult) primary stromal† Pdgfrα+ Sca1- | |
| 5 | BM MPC’s (Pdgfrα+ Sca1+) | GSM437799 | BM (adult) primary stromal† Mesenchymal progenitor cells (MPC) (Pdgfrα+ Sca1+) | |
| 6 | BM MPC’s (Alcam+ Sca1+) | GSM437796 | BM (adult) primary stromal† Mesenchymal progenitor cells (MPC) (Alcam+ Sca1+) | |
| 7 | BM MSC-like derived clones | GSM795638-40 | BM MSC-derived cell line, Wnt1Cre/R26R β-gal- selected clone passage>10 | § |
| 8 | BM NCSC-derived clones | GSM795641-43 | BM NCSC-derived cell line, Wnt1Cre/R26R β-gal- selected clone passage>10 | |
| 9 | E13.5 vascular segment (mesoderm-derived) | GSM261911 | E13.5 internal carotid artery vascular segment (smooth muscle mesoderm-derived) | ¶ |
| 10 | E13.5 vascular segment (NC derived) | GSM261912 | E13.5 external carotid artery vascular segment (smooth muscle NC-derived) | |
| 11 | BM (adult) Nes-GFP+ | GSM545815-17 | BM (adult) primary (CD45-) Nes-GFP+ cells | ** |
| 12 | BM (adult) Scf-GFP+ | GSM821066-68 | BM (adult) primary Scf-GFP+ cells | †† |
| 13 | P0 Schwann cells | GSM15386-88 | P0 primary Schwann cells (Plp-GFP+) from sciatic nerve | ‡‡ |
| 14 | E12.5 Schwann cell precursors (SCPs) | GSM15373-75 | E12.5 primary Schwann cell precursors (Plp-GFP+) from sciatic nerve | |
| 15 | E14.5 Enteric Neural crest cells (ENCC) | GSM844492-94 | E14.5 primary ENCCs (Wnt1Cre/R26-YFP+) from gut | |
| 16 | E9.5 Neural crest stem cells (NCSC) | GSM15370-72 | E9.5 trunk primary Plp-GFP+ cells (migrating NCSCs) | |
| 17 | E18.5 Schwann cells | GSM15383-85 | E18.5 primary Schwann cells (Plp-GFP+) cells from sciatic nerve |
-
*
Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/)
-
†
CD45-CD31-Ter119-
-
References:
-
‡
Nakamura et al. Isolation and characterization of endosteal niche cell populations that regulate hematopoietic stem cells. Blood (2010) vol. 116 (9) pp. 1422-32.
-
§
Wislet-Gendebien et al. Mesenchymal stem cells and neural crest stem cells from adult bone marrow: characterization of their surprising similarities and differences. Cell Mol Life Sci (2012)vol. 69 (15) pp. 2593-608.
-
¶
Zhang et al. Origin-specific epigenetic program correlates with vascular bed-specific differences in Rgs5 expression. FASEB J (2012) vol. 26 (1) pp. 181-91.
-
**
Méndez-Ferrer et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature (2010) vol. 466 (7308) pp. 829-34.
-
††
Ding and Morrison. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature (2013) pp. 1-6.
-
‡‡
Buchstaller et al. Efficient isolation and gene expression profiling of small numbers of neural crest stem cells and developing Schwann cells. J Neurosci (2004) vol. 24 (10) pp. 2357-65.
Additional files
-
Supplementary file 1
Summary of mouse strains used in this study.
- https://doi.org/10.7554/eLife.03696.019