Dominant drug targets suppress the emergence of antiviral resistance
Figures
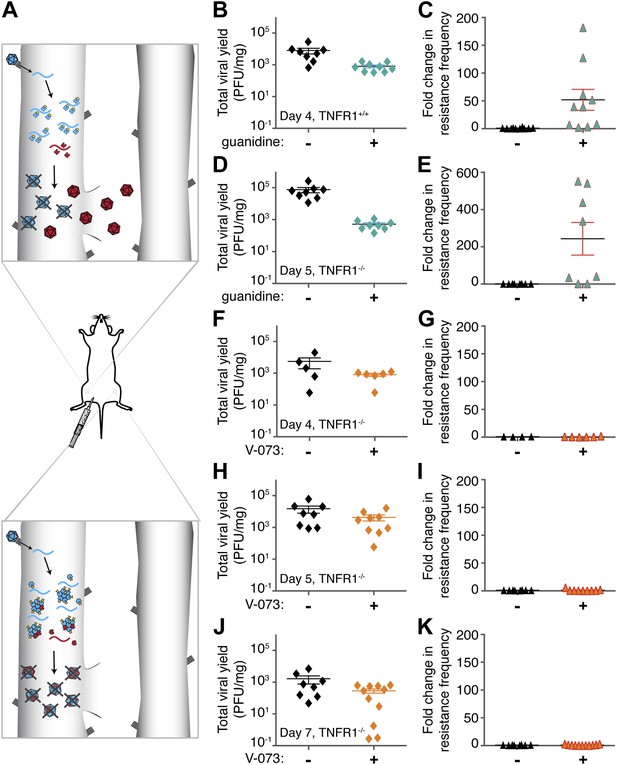
Emergence of drug-resistant variants in non-dominant and dominant drug targets in mice.
(A) Representation of drug-resistant viral amplification when a non-dominant drug target is inhibited (top) but its inhibition by drug-susceptible variants when a dominant drug target, such as an oligomeric structure, is inhibited (bottom). See text for discussion. (B–K) Tnfr1+/+ (B, C) or Tnfr1−/− (d-k) PVR-expressing mice were infected with 1 × 107 PFU Mahoney type 1 poliovirus by intramuscular inoculation and treated with 76 mg/kg/day guanidine (B–E, green symbols) or 10 mg/kg/day V-073 (1-[(2-chloro-4-methoxyphenoxy)methyl]-4-[(2,6-dichlorophenoxy)methyl]benzene) (F–K, orange symbols). Inoculated muscles were harvested at times indicated. Total viral yields (PFU/mg tissue) in muscle samples of mice treated with guanidine (B, D), V-073 (F, H, J) or vehicles (black) are shown. Fold changes in the frequency of drug-resistant variants in mice treated with guanidine (C, E) or V-073 (G, I, K) are shown. Fold change calculated by dividing sample value by mean value of control mice. p-values: 0.02 (B), 0.003 (C), 0.0002 (D) 0.02 (E), and 0.01 (F, H, J combined).
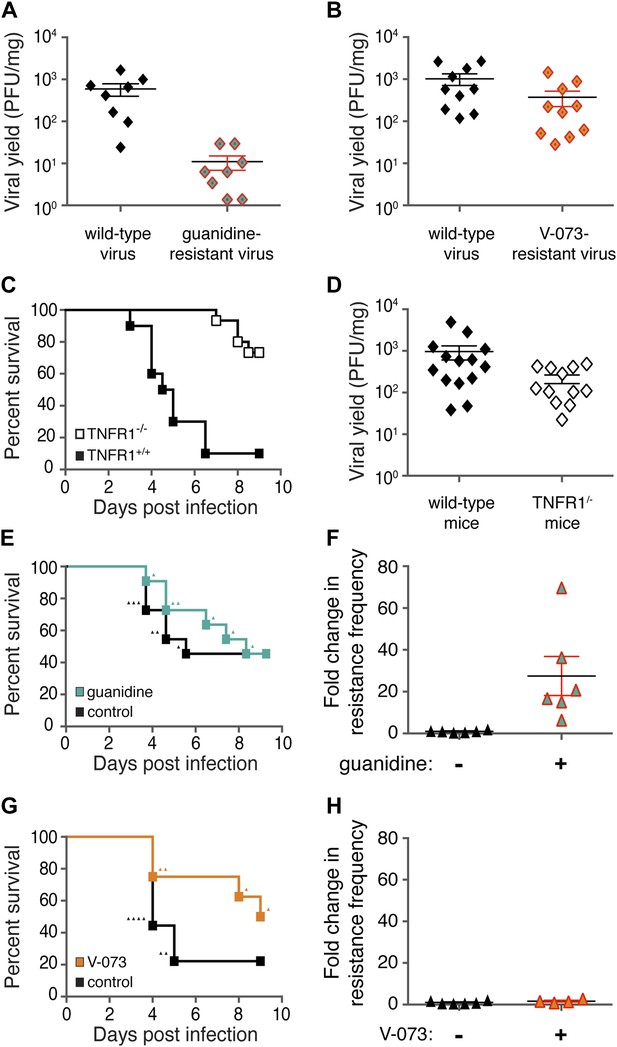
Differences in frequencies of guanidine-resistant and V-073-resistant viruses do not result from differences in viral fitness or murine health.
(A, B) Total viral yield (PFU/mg) of mouse-adapted wild-type and guanidine-resistant virus (p = 0.0006) or wild-type and V-073-resistant virus in muscles of Tnfr1−/− PVR+/+ mice 4 days post infection. (C) Survival of Tnfr1+/+ and Tnfr1−/− mice after infection. Tnfr1+/+, n = 10; Tnfr1−/− mice, n = 15. p = <0.0001. (D) Viral yield (PFU/mg) in muscles of Tnfr1−/− mice compared to Tnfr1+/+ mice 4 days post infection. p = 0.035. (E, F) Survival (E) and frequency of drug resistance (F) of cPVR mice in the presence and absence of treatment with 76 mg/kg/day guanidine. For both groups in survival analysis, n = 11. Triangles in (E) indicate individual mice that are analyzed in (F). (G, H) Survival (G) and frequency of drug resistance (H) of cPVR mice in the presence and absence of treatment with 3 mg/kg/day V-073. For survival analysis, untreated mice, n = 9; treated mice n = 8. Triangles in (G) indicate individual mice that are analyzed in (H). Mouse tissue was harvested immediately upon the first observation of paralysis (E–H). All mice were inoculated with 1 × 106 PFU virus.
Co-infection experiments to test genetic dominance of drug-susceptible genomes.
Differential growth of defined viral variants resistant to compounds inhibitory to three different viral targets in mixed infections. (A) Chemical structure of guanidine, which inhibits the NTPase activity of poliovirus 2C protein. (B) A previously characterized guanidine-resistant poliovirus variant (2C-N179G) was genetically reconstructed. The yield of 2C-N179G virus at (C) MOI of 10 PFU/cell and (D) MOI of 0.1 PFU/cell in the presence of guanidine and increasing amounts of wild-type, drug-susceptible virus in the same cells was determined. (E) Chemical structure of V-073, which inhibits the function of the icosahedral poliovirus capsid (F) A V-073-resistant poliovirus variant (VP3-A24V) was identified and genetically reconstructed in the Mahoney type 1 viral background. The yield of VP3-A24V virus in the (G) presence and (H) absence of V-073 and increasing amounts of wild-type, drug-susceptible virus in the same cells was determined. (I) V-073-resistant variant VP3-A24V and (J) VP1-I94F in the epidemic Hispaniola strain of poliovirus were identified and reconstructed. The yields of (K) VP3-A24V and (L) VP1-I94F Hispaniola type 1 poliovirus in the presence of V-073 and increasing amounts of wild-type, drug-susceptible Hispaniola virus in the same cells were determined. (M) Chemical structure of rupintrivir (trans-(4S,2R,5S,3S)-4-{2-4-(4-fluorobenzyl)-6-methyl-5’-[(5-methylisoxazole)-3-carbonylamino]-4-oxoheptanoylamino}-5-(2-oxopyrrolidin-3-yl)pent-2-enoic acid ethyl ester), which targets the active site of poliovirus 3C proteinase. (N) A rupintrivir-resistant poliovirus variant (3C-G128Q) was identified and genetically reconstructed. The yield of 3C-G128Q virus at (O) an MOI of 10 PFU/cell and at (P) 0.1 PFU/cell in the presence of rupintrivir and increasing amounts of wild-type, drug-susceptible virus in the same cells was determined. *p < 0.01.
V-073-susceptible and V-073-resistant virus form mixed capsids during co-infection.
(A, B) Depiction of ‘chimeric’ (A) and ‘pooled’ (B) viral preparations. Red subunits represent V-073-resistant, FLAG-tagged capsids, and grey subunits represent V-073-susceptible, HA-tagged capsids. Anti-FLAG antibodies are represented in pink. (C) Construction of FLAG- and HA-tagged virus. Sequence of FLAG and HA peptides inserted into antigenic site 1 in the poliovirus genome, replacing amino acids 96-101 and 97-102 of VP1, respectively. (D) Viral yield (PFU/ml) of the wild-type, FLAG-tagged and HA-tagged poliovirus variants in the absence or presence of V-073 over a single infectious cycle at an MOI of 10. Data represent means ± s.t.d. of two replicates. (E) Fraction of capsids with V-073-resistant phenotype in anti-FLAG immunoprecipitates as labeled. HA-tagged sample was below the limit of detection. Data represent means ± s.t.d. of two replicates. (F) Immunoblots of VP1 proteins collected by anti-FLAG immunoprecipitation of cell lysates prepared from infections indicated. ‘Pooled’ sample contains lysates from independently grown FLAG- and HA-tagged viruses, whereas ‘co-infected’ sample was obtained from cells infected with both FLAG- and HA-tagged variants. All variants were infected at an MOI of 10 PFU/cell.
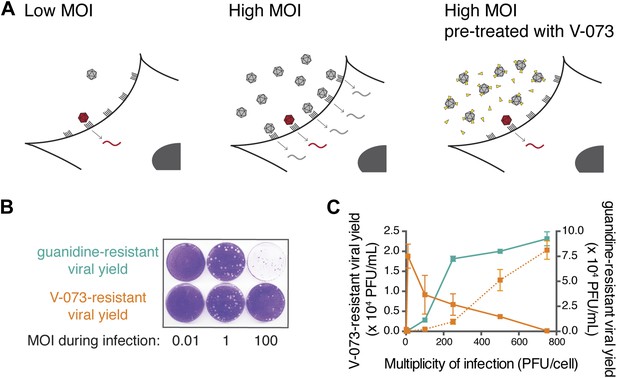
Assaying the dominance of drug-susceptible genomes via the MOI-dependent detection of drug-resistance.
(A) Cartoon of proposed infection at low MOI (left) and high MOI without V-073 pretreatment (middle) or with V-073 pretreatment (right). Grey represents drug-susceptible variants, red represents drug-resistant variant and drug is depicted as yellow triangles. (B) Drug-resistant virus plaques following infections with wild-type Hispaniola strain at increasing MOIs in the presence of guanidine or V-073. (C) Drug-resistant viral yield (PFU/ml) in presence of guanidine (green line) or V-073 without pretreatment of viral stock (solid orange line) or with pretreatment (dotted orange line).





