The glucuronyltransferase B4GAT1 is required for initiation of LARGE-mediated α-dystroglycan functional glycosylation
Figures
Postulated α-DG modifying enzymes are involved in post-phosphorylation processes in the Golgi prior to LARGE.
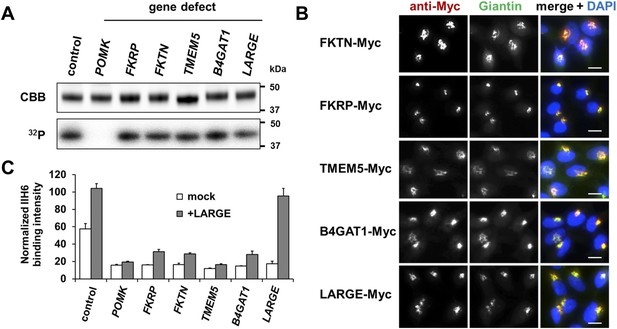
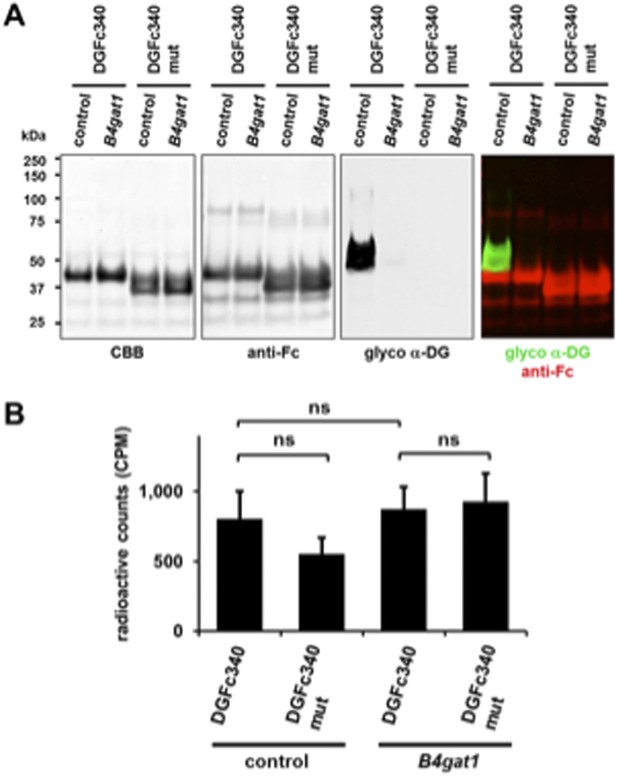
(A) Phosphorylation of Fc-tagged DGFc340 in the context of α-DG glycosylation defects. Fc-tagged DGFc340 was produced in [32P] orthophosphate-labeled fibroblasts from control and glycosylation-deficient patients and mice (Table 1). The DGFc340 was isolated from the culture medium by using protein A-agarose and the samples were separated by SDS PAGE. Gels were stained with Coomassie brilliant blue (CBB) and analyzed by phosphorimaging (32P). (B) Subcellular localization of α-DG modifying putative glycosyltransferases, as assessed by immunofluorescence. HEK293T cells stably expressing c-Myc-tagged proteins were stained with anti-Myc (red), anti-Giantin (Golgi marker, green) and 4 ́,6-diamidino-2-phenylindole (DAPI, nuclei, blue). Individual stainings for c-Myc and Giantin are shown in greyscale and a merged image is shown in color. Scale bars indicate 10 µm. (C) Quantitative On-Cell protein blot analysis of LARGE-induced α-DG glycosylation hyperglycosylation in glycosylation-deficient cells. α-DG glycosylation status was tested with and without forced LARGE overexpression by adenovirus mediated gene transfer. The On-Cell Western blots were probed with an antibody against the glycosylated form of α-DG (IIH6). IIH6 On-Cell quantitative data were normalized with DRAQ5 cell DNA dye (n = 3). Error bars, SD.
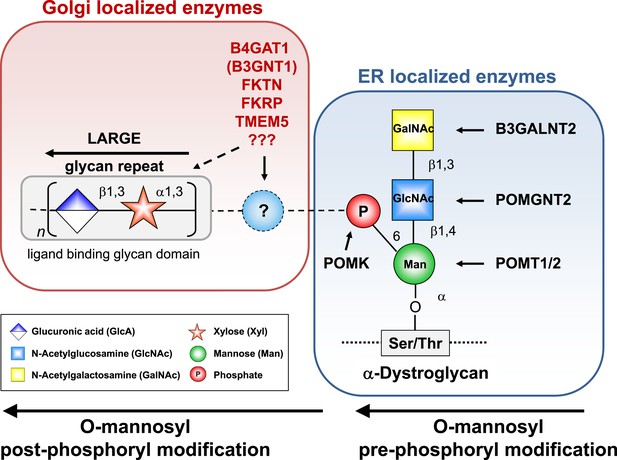
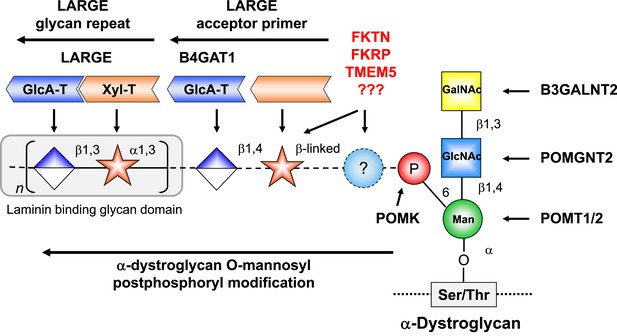
α-DG functional glycosylation and known proteins contributing to its synthesis.
α-DG core M3 functional glycosylation can be divided in 2 major processing steps. O-mannosyl pre-phosphoryl modification which is carried out by enzymes in the endoplasmic reticulum (ER) (highlighted in the blue box) and O-mannosyl post-phosphoryl modification by known or putative glycosyltransferases in the Golgi (highlighted in the red box). Both gene products with known function (black) and gene products with currently unidentified function (red) are indicated. The putative glycosyltransferases B4GAT1 (B3GNT1), FKTN, FKRP and TMEM5 are proposed to act prior to LARGE which adds a GlcA-Xyl heteropolymer that is responsible for ligand binding. However based on current knowledge it cannot be completely ruled out that they are involved in the modification of the LARGE glycan repeat itself to modulate ligand binding.
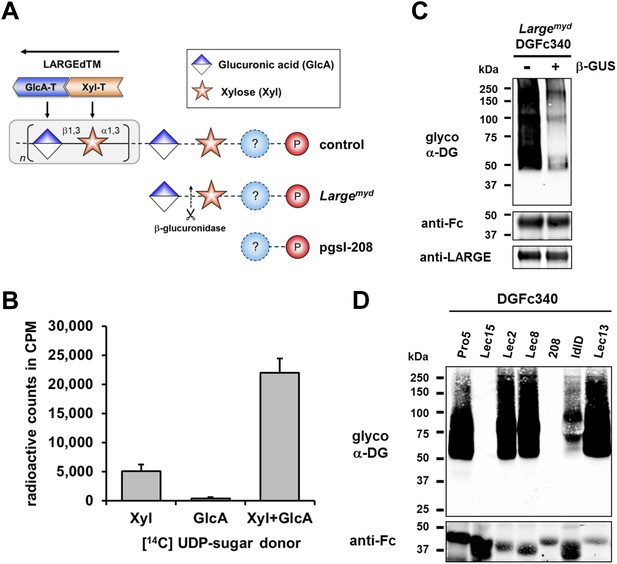
β-GlcA serves as an acceptor sugar for LARGE modification starting with xylose.
(A) Schematic diagram showing the α-DG post-phosphoryl modification in the context of control and glycosylation defects. LARGE adds the ligand-binding glycan to α-DG via a proposed glucuronic acid (GlcA) acceptor. LARGEdTM catalytic domains Xyl-T (orange) and GlcA-T (blue) are highlighted in color. Depicted are also the hypothesized terminal sugar structures of glycosylation-deficient cell lines Largemyd (Large-deficient) and pgsI-208 (UDP-xylose deficient). Cleavage of terminal β-GlcA by exoglycosidase β-glucuronidase (β-GUS) in Largemyd is indicated (scissor symbol). (B) Transfer of [14C] radiolabeled Xyl and GlcA to DGFc340 by LARGEdTM. Fc-tagged DGFc340 was produced in Largemyd (Large-deficient) MEF cells and isolated from the culture medium using protein A-agarose. The protein A-bound DGFc340 was used as acceptor in a LARGEdTM reaction with radiolabeled [14C] UDP-Xyl and/or [14C] UDP-GlcA sugar donors. The figure represents the transfer of radiolabeled saccharides onto the donor DGFc340 (n = 3). Error bars represent SD (C) β-Glucuronidase pre-treatment of DGFc340 from Largemyd deficient cells impairs LARGEdTM modification. Protein A-bound DGFc340 (acceptor) isolated from transfected Largemyd MEFs was digested with β-glucuronidase (β-GUS) prior to the LARGEdTM (enzyme) reaction, which included UDP-Xyl and UDP-GlcA as sugar (donors). After incubation with LARGEdTM DGFc340 (acceptor protein) was subjected to protein blotting with antibodies against the glycosylated form of α-DG (IIH6), against Fc and against LARGE (Rb331). (D) The ability of LARGEdTM to modify DGFc340 is impaired in the context of sugar donor-deficient CHO mutant cell lines. Fc-tagged DGFc340 was produced in various glycosylation-deficient Lec CHO cells and isolated from the culture medium using protein A-agarose. As in (C) protein A-bound DGFc340 acceptor was used in a LARGEdTM reaction and analyzed by protein blotting.
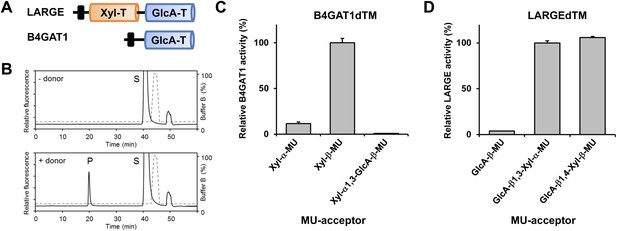
B4GAT1 has xylose β1,4 glucuronyltransferase activity.
(A) Schematic representation of LARGE and B4GAT1 functional domains. GlcA-T (blue), Xyl-T (orange) and transmembrane domain (black) are indicated. (B) Representative HPLC profiles of the reaction product generated in the absence (top) and presence (bottom) of a UDP-GlcA sugar (donor) in a reaction mix containing Xyl-β-MU (acceptor) and B4GAT1dTM (enzyme). Samples were separated on an LC-18 column. P, product. S, unreacted substrate. Dotted line, %B buffer. (C) Comparison of B4GAT1dTM GlcA-T activity with respect to various xylose-MU acceptor sugars. Relative activity (%) with respect to Xyl-β-MU acceptor (specific activity: 0.2 µmol/h/mg) is shown (n = 3). Error bars represent SD. (D) Comparison of LARGEdTM Xyl-T activity with respect to various monosaccharide and disaccharide GlcA-MU acceptor sugars. Relative activity (%) with respect to intrinsic LARGE polymer specific activity with GlcA-β1,3-Xyl-α-MU disaccharide acceptor (0.08 µmol/h/mg) (n = 3). Error bars represent SD.
-
Figure 3—source data 1
Chemical shifts (ppm) of the signals in the 1H and 13C NMR spectra of the enzymatic reaction product of GlcA-β1,4-Xyl-β-MU of the glycosyltransferase B4GAT1.
- https://doi.org/10.7554/eLife.03941.008
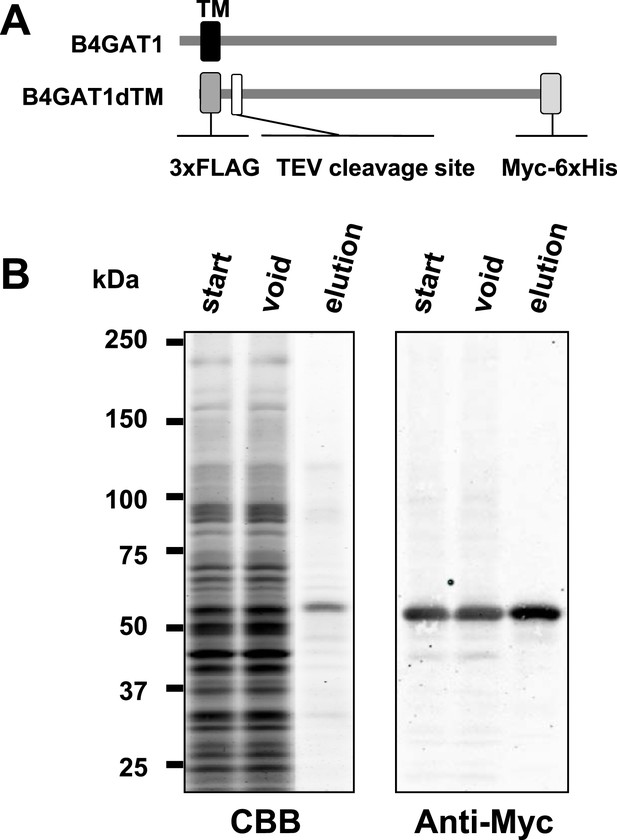
Purification of B4GAT1dTM.
(A) Schematic representation of B4GAT1 and the B4GAT1dTM construct used in the enzymatic activity assay. The transmembrane (TM) sequence was replaced with a 3xFLAG-TEV tag sequence and the C-terminus was modified with a Myc-6xHis-tag. (B) Purification of recombinant B4GAT1dTM from bioreactor culture medium. The recombinant protein was expressed in HEK293T cells and purified from the culture medium using Talon metal-affinity resin. The bioreactor medium samples before (start) and after the purification (void) as well as the eluted purified protein (elution) were analyzed by immunoblotting with anti-Myc (4A6) antibody. CBB, stained with Coomassie Brilliant blue.
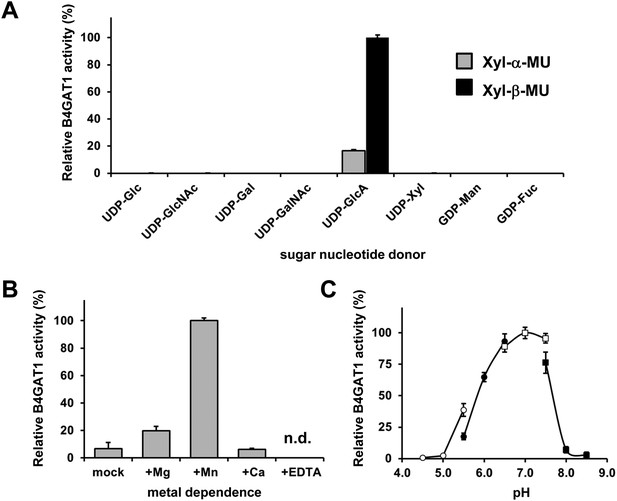
Basic characterization of the xylose β1,4-glucuronyltransferase activity of B4GAT1.
(A) Donor sugar specificity of B4GAT1dTM. Representative data from two independent assays, demonstrating relative activity (%) of B4GAT1dTM (enzyme) GlcA-T toward Xyl-α-MU and Xyl-β-MU (acceptor) when tested with various sugar nucleotides (donor). The specific activity set as 100% for acceptor Xyl-β-MU was 0.2 µmol/hr/mg. No sugar other than GlcA was transferred to the acceptors to a significant extent. (B) Metal dependence of the B4GAT1dTM GlcA-T activity. Activity assay was carried out in the presence or absence of each metal ion or EDTA (10 mM), and results are shown as relative activity (%). The GlcA-T activity in the presence of Mn2+ (specific activity: 0.26 µmol/h/mg) was arbitrarily set at 100% (n = 3). n.d., not detected. Error bars represent SD (C) pH optimum of B4GAT1dTM GlcA-T activity. Data from three independent experiments are shown as relative activity (%). The highest activity (specific activity: 0.22 µmol/h/mg) in the dataset was arbitrarily set at 100%. The GlcA-T assays were carried out using Xyl-β-MU as acceptor. The buffers used were: acetate for pH 4.5––5.5 (open circle), MES for pH 5.5––6.5 (closed circle), MOPS for pH 6.5––7.5 (open square) and Tris–HCl for pH 7.5––8.5 (closed square). The details of the conditions are presented in the Materials and methods section. Error bars represent SD.
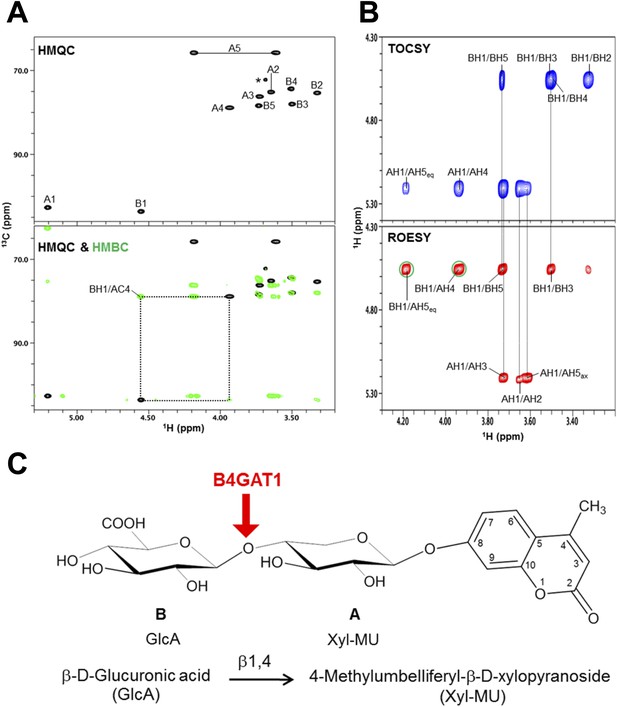
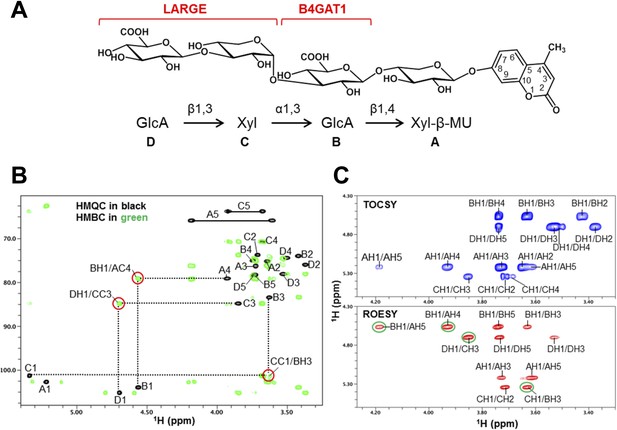
NMR analysis reveals that B4GAT1 is a β1,4 glucuronyltransferase.
(A) HMQC spectrum (top) and overlay of HMQC (black) and HMBC (green) spectra (bottom) for the B4GAT1 enzymatic reaction product. The cross-peaks are labeled with a first letter representing the subunit designated in C and the rest of the label representing the position on that subunit. The observed interglycosidic cross-peak BH1/AC4 in the HMBC spectrum clearly demonstrates the presence of a 1→4 interglycosidic linkage between the residues B and A. The cross-peak marked with a star represents an impurity. (B) TOCSY (top) and ROESY (bottom) spectra of the B4GAT1 enzymatic reaction product collected with a mixing time of 77 and 300 ms, respectively. The cross-peaks are labeled as in (A). The observed interglycosidic ROEs are indicated in green circles. The ROE data indicate that both residues exist in β-configurations. (C) Schematic depiction of the disaccharide structure produced by B4GAT1, with the sugar units labeled A and B.
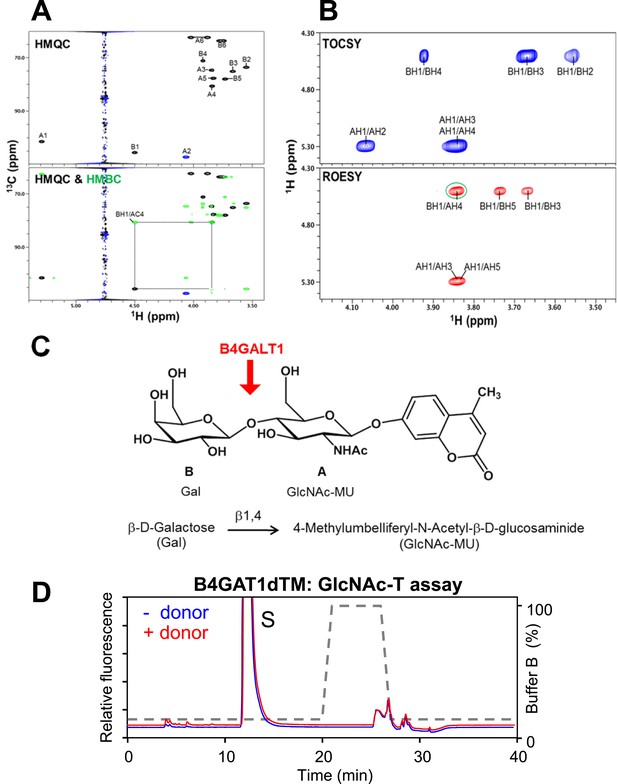
Test B4GAT1 for GlcNAc transferase activity with iGnT substrate Gal-β1,4-GlcNAc-β-MU.
(A) Using B4GALT1 we synthesized the hypothesized iGnT substrate Gal-β1,4-GlcNAc-β-MU by transferring a β1,4 Galactose to the acceptor GlcNAc-β-MU. The purified Gal-β1,4-GlcNAc-β-MU disaccharide was further analyzed by NMR. (B) HMQC spectrum. The folded peak is shown in blue. The cross peaks are labeled with a first letter representing the subunit as designated in the structure shown above the spectra and the rest of the label representing the position on that subunit. Overlay of HMQC (black and blue) and HMBC (green) spectra. The strong cross peak labeled as BH1/AC4 was detected in the HMBC spectrum, demonstrating the presence of a 1→4 interglycosidic linkage between Gal and GlcNAc. (C) TOCSY spectrum collected with a mixing time of 77 ms. ROESY spectrum collected with a mixing time of 300 ms. The observed strong NOEs from BH1 to BH3 and BH5 (cross peaks labeled as BH1/BH3 and BH1/BH5) demonstrate that the Gal has a β-configuration. Similarly, the observed strong NOEs from AH1 to AH3 and AH5 (cross peaks labeled as AH1/AH3 and AH1/AH5) demonstrate that the GlcNAc has a β-configuration. A strong interglycosidic NOE was observed between BH1 and AH4 (green circle), which is consistent with the 1 to 4 linkage as determined from the HMBC spectrum. (D) Representative HPLC profiles of the reaction product generated in the absence (blue) and presence (red) of a UDP-GlcNAc sugar (donor) in a reaction mix containing Gal-β1,4-GlcNAc-β-MU (acceptor) and B4GAT1dTM (enzyme). Samples were separated on an LC-18 column. S, unreacted substrate. Dotted line, %B buffer.
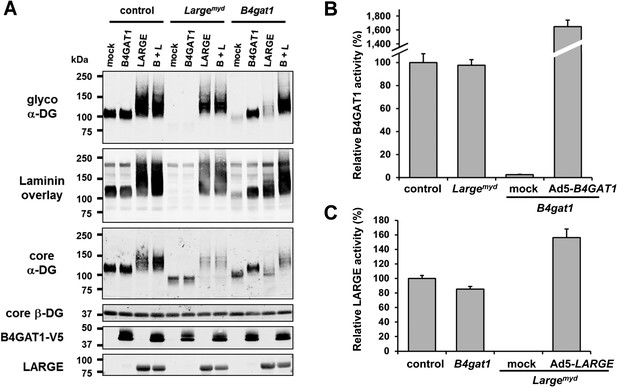
B4gat1-deficient MEFs have impaired α-DG functional glycosylation and endogenous B4GAT1 activity.
(A) Functional glycosylation and complementation analysis of α-DG in wild-type, Large- and B4gat1-deficient MEFs. Immunoblots and laminin overlay assay of WGA-enriched cell lysates extracted from WT, Largemyd (Large-deficient) and B4gat1-deficient MEFs . As indicated MEFs were uninfected (mock) or infected with adenovirus constructs expressing B4GAT1, LARGE or both (B + L). Antibodies used were: glyco α-DG (IIH6), core α-DG, core β-DG (AP83), anti-V5 and anti-LARGE (Rb331). (B) Comparison of endogenous B4GAT1 GlcA-T activity in control, Large- and B4gat1-deficient MEFs. Additionally, B4gat1-deficient MEFs (B4gat1LacZ/M155T) complemented with control B4GAT1 expressing adenovirus (Ad5) were tested. Cell lysates were used as enzyme source to measure endogenous B4GAT1 activity. Relative activity (%) with respect to control MEFs specific activity (91.6 pmol/h/mg) is shown (n = 3). Error bars represent SD. (C) Comparison of endogenous LARGE GlcA-T activity in control, Large- and B4gat1-deficient MEFs. Additionally, Large-deficient MEFs complemented with control LARGE expressing adenovirus (Ad5) were tested. WGA enriched glycoprotein samples were used as enzyme source to measure endogenous LARGE activity. Relative activity (%) with respect to control MEFs specific activity (0.52 pmol/h/mg) is shown (n = 3). Error bars represent SD.
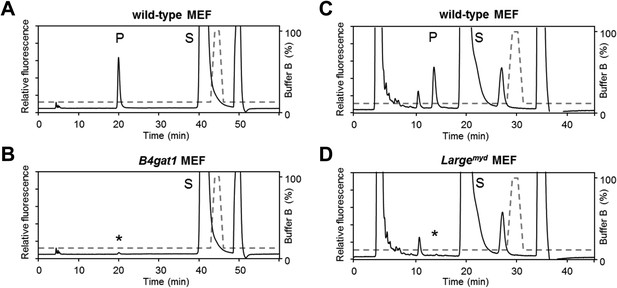
B4gat1-deficient MEFs have impaired endogenous B4GAT1 activity.
Representative HPLC profiles of the reaction product are shown. (A/B) Endogenous B4GAT1 enzyme activity of cell lysates from wild-type (A) and B4gat1-deficient (B) MEFs . B4gat1-deficient MEFs (B4gat1LacZ/M155T) show some residual activity <3% (asterisk). (C/D) Endogenous LARGE enzyme activity of WGA-enriched cell lysates from control (C) and Largemyd (D) (Large-deficient) MEFs. Largemyd MEFs lack LARGE activity and do not show any residual activity (asterisk). The details of the conditions are provided in the Materials and methods section.
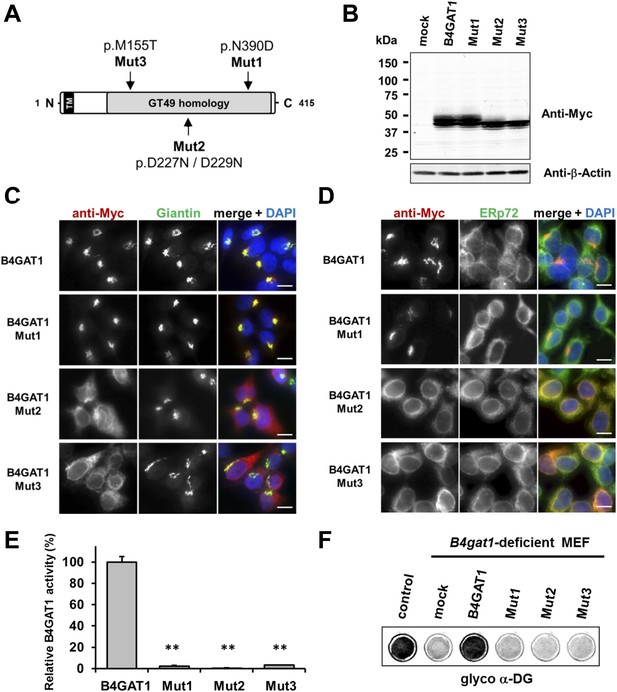
Expression analysis and GlcA-T enzyme activity of B4GAT1 mutant constructs.
(A) Schematic presentation shows B4GAT1 enzyme product with functional domains and B4GAT1 mutations Mut1-Mut3 are indicated. (B) Expression analysis of B4GAT1-Myc control and mutant constructs in stable HEK293T cells. Immunoblotting of cell lysates from HEK293T cells stably overexpressing wild-type B4GAT1-Myc and mutant constructs (Mut1, Mut2 and Mut3) with anti-Myc antibody and β-Actin (loading control). (C/D) Subcellular localization of B4GAT1-Myc control and mutant constructs in stable HEK293T cells (see B). B4GAT1-Myc constructs were stained with anti-Myc (red), (C) anti-Giantin (Golgi marker, green), (D) anti-ERp72 (ER marker, green) and 4 ́,6-diamidino-2-phenylindole (DAPI, nuclei, blue). Individual stainings for c-Myc Giantin and ERp72 are shown in greyscale, and merged images are shown in color. Scale bars indicate 10 µm. (E) B4GAT1 enzyme activity in cell lysates from stable HEK293T cells overexpressing B4GAT1-Myc wild-type and B4GAT1-Myc mutant constructs (Mut1-Mut3). Relative activity (%) with respect to B4GAT1wild-type specific activity (19.8 nmol/hr/mg) is shown (n = 3). Error bars represent SD, Statistical analyses were performed by two-tail Student's t test. **p < 0.001. (F) Complementation of B4gat1-deficient (B4gat1LacZ/M155T) MEF cells with B4GAT1-Myc control and mutant constructs. B4gat1-deficient MEFs were nucleofected with a wild-type or mutant B4GAT1 expression construct. α-DG functional glycosylation was analyzed by On-Cell-Western analysis. α-DG functional glycosylation was detected with α-DG glyco (IIH6) antibody.
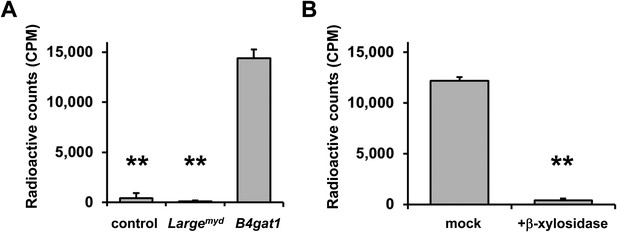
β-xylose is the endogenous acceptor for B4GAT1.
(A) B4GAT1dTM enzymatic transfer of [14C] radiolabeled GlcA to DGFc340. Fc-tagged DGFc340 (acceptor) was produced in control, Largemyd (Large-deficient) and B4gat1-deficient MEFs and isolated from the culture medium using protein A-agarose. The protein A-bound Fc340 was used as acceptor in a B4GAT1dTM (enzyme) reaction with radiolabeled [14C] UDP-GlcA sugar (donor). The figure represents the transfer of radiolabeled GlcA onto the donor DGFc340 (n = 3). Error bars represent SD. Statistical analyses were performed by two-tail Student's t test. **p < 0.001. (B) β-Xylosidase pre-treatment impairs B4GAT1dTM transfer of [14C] radiolabeled GlcA. DGFc340 (acceptor) from B4gat1-deficient MEFs was digested with β-xylosidase prior to the B4GAT1dTM (enzyme) transfer reaction with [14C] UDP-GlcA sugar (donor). The figure represents the transfer of radiolabeled GlcA onto the donor DGFc340 (n = 3). Error bars represent SD Statistical analyses were performed by two-tail Student's t test. **p < 0.001.
NMR analyses of the tetrasacharide generated from B4GAT1dTM and LARGEdTM enzymatic reactions.
(A) Schematic depiction of the tetrasaccharide structure produced by the sequential reactions of B4GAT1dTM followed by LARGEdTM with the sugar units labeled A-D to indicate the order of their addition. (B) Overlay of the HMQC (black) and HMBC (green) spectra of the tetrasaccharide. All cross-peaks in the HMQC spectrum are labeled. Three interglycosidic cross-peaks detected in the HMBC spectrum are also labeled and indicated with red circles. The peaks are labeled with a first letter representing the subunit designated in A, and the rest of the label representing the position on that subunit. (C) TOCSY spectrum (top) and ROESY spectrum (bottom) of the tetrasaccharide. The TOCSY and ROESY spectra were collected with mixing time of 77 and 300 ms, respectively. The cross-peaks are labeled as in B. The observed interglycosidic ROEs are indicated with green circles.
-
Figure 7—source data 1
Chemical shifts (ppm) of the signals in the 1H and 13C NMR spectra of the tetrasaccharide of GlcA-β1,3-Xyl-α1,3-GlcA-β1,4-Xyl-β-MU produced by B4GAT1 and LARGE.
- https://doi.org/10.7554/eLife.03941.018
Model of proposed α-DG O-mannosyl laminin-binding glycan structure and the enzymes that contribute to its synthesis.
Post-phosphoryl modification of α-DG requires B4GAT1 (β1,4 glucuronyltransferase); this enzyme generates the acceptor glycan, which serves as a primer for the glycosyltransferase LARGE to initiate synthesis of the laminin-binding glycan. Both gene products with known function (black) and gene products with currently unidentified function (red) are indicated.
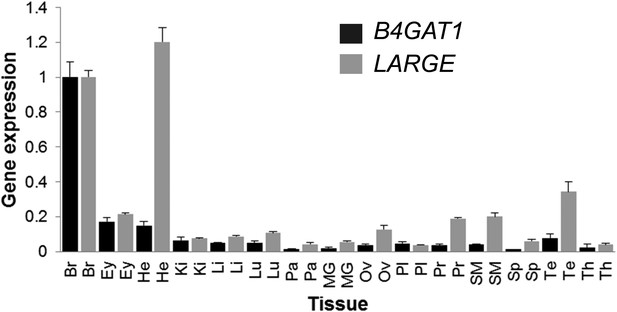
B4GAT1 and LARGE expression in human tissues.
qPCR revealing ubiquitous B4GAT1 and LARGE expression in all tissues analyzed, with highest expression of LARGE in brain and heart. cDNA was synthesized using random primers and oligo(dT) on commercially available human tissues RNAs. For each tissue, B4GAT1 and LARGE were specifically amplified, in triplicate, in the presence of SYBRgreen, and their expressions was normalized to that of the 28S RNA (normalization control). The expression in each tissue is referenced with respect to that in brain. Analyzed tissues: Br (brain), Ey (eye), He (heart), Ki (kidney), Li (liver), Lu (lung), Pa (pancreas), MG (mammary gland), Ov (ovary), Pl (placenta), Pr (prostate), SM (skeletal muscle), Sp (spleen), Te (testis), Th (thymus). Error bars represent.
[3H] Xylose metabolic labeling of DGFc340. DGFc340 and DGFc340mut fusion proteins were enriched by Protein A-agarose from culture medium of [3H] Xylose labeled control and B4gat-deficient MEF cells. (A) Expression and functional glycosylation analysis of Fc fusion proteins by Coomassie brilliant blue (CBB) stain and by immunoblot. The SDS-PAGE was stained with CBB and the immunoblot was incubated with antibodies against glyco α-DG (IIH6) and anti-Fc. (B) The figure represents the incorporation of radiolabeled [3H] Xylose into DGFc340/DGFc340mut fusion proteins (n = 3). Error bars represent s.d. Statistical analyses were performed by two-tail Student's t test. ns: not significant (p > 0.05).
Tables
Summary of features of control and glycosylation-deficient cell lines
| Mutant gene | Clinical phenotype | Cell type | Nucleotide variant | Amino acid | reference |
|---|---|---|---|---|---|
| Control (human) | none | Human skin fibroblast | CRL-2127 (ATCC) | ||
| POMK | WWS/MEB | Human skin fibroblast | 14bp homozygous deletion (c.720_733delGCTGGTG AGTGCG)], homozygous | p.Leu241Profs*26 | (Yoshida-Moriguchi et al., 2013) |
| FKTN | WWS | Human skin fibroblast | c.385delA | p.I129fsX1 | GM16192 |
| c.1176C > A, heterozygous | p.Y392X | (Coriell Cell Repository) | |||
| FKRP | WWS | Human skin fibroblast | c.1A > G, homozygous | p.M1V | (Van Reeuwijk et al., 2010) |
| TMEM5 | WWS | Human skin fibroblast | c.1101 G > A, homozygous | p.G333R | unpublished |
| B4gat1 (B3gnt1) | CMD | MEF | c.464T > C, compound het with LacZ null allele, B4gat1LacZ/M155T | p.M155T | (Wright et al., 2012) |
| Largemyd | CMD | MEF | deletion of exons 5−7, homozygous | (Grewal et al., 2001) | |
| Control (mouse) | none | MEF |





