Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems
Figures
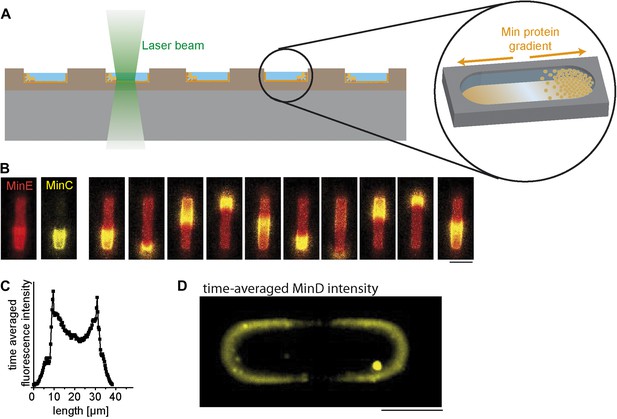
Protein gradients self-organize in soft-polymer containers.
(A) Experimental setup. Purified proteins of the MinC/D/E system were reconstituted in membrane-clad soft-polymer compartments and imaged by confocal microscopy. Profiles and electron micrographs of the compartments are shown in Figure 1—figure supplement 1 and a more detailed description of the assay to reconstitute Min protein oscillations is presented in Figure 1—figure supplement 2. Figure 1—figure supplement 3 demonstrates how the intensity profiles of MinD and MinE along the length axis of the compartments are modulated with time. To mimic a bacterial membrane we used E. coli polar lipids to generate supported lipid membranes. A more detailed description of how lipid composition influences pattern formation of Min proteins is presented in Figure 1—figure supplement 4. (B) In cell-shaped compartments, eGFP-MinC (yellow) follows the oscillations of MinD and MinE (red). Comparable results were obtained in more than a hundred soft-polymer compartments. Confocal time-lapse images with the image plane at the bottom of the compartment. Protein concentrations: 1 µM MinD, 0.9 µM MinE, 0.1 µM MinE.Atto655 (red), 0.05 µM eGFP-MinC (yellow). Time between individual frames: 30 s. Scale bar: 5 µm. (C) The time-averaged concentration profile of MinC along the long axis of a compartment has a distinct concentration minimum in the middle of the compartment. The time averaged distribution of protein concentrations was calculated by acquiring time-lapse-images with the focal plane at the middle of the compartment and averaging the intensity of the acquired frames. (D) Time-averaged fluorescent signal of eGFP-MinD with image plane in the middle of a compartment. An intensity offset is subtracted to better visualize the gradient along the boundary of the compartment. 0.9 µM MinD, 0.1 µM eGFP-MinD, 1 µM MinE. Scale bar: 5 µm. Stable pole-to-pole oscillations which result in the time-averaged gradient of MinD are severely affected if the membrane targeting sequence of MinE is deleted (Figure 1—figure supplement 5).
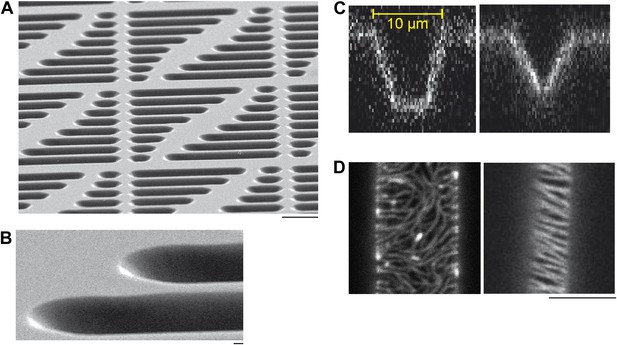
Micro compartments in a soft-polymer chip.
(A) Scanning electron micrographs of a multi-compartment PDMS chip. Arrays of compartments with different geometries, for example, different lengths, provide the possibility to study geometry dependent effects of protein self-organization on one chip. Each compartment serves as reaction container to reconstitute protein networks. Scale bar: 20 µm. (Note that the scale bars in the electron micrographs only applies to the x-direction of the picture. The sample was tilted in the other direction to create the three-dimensional impression of electron micrographs.) (B) Same sample as descibed in (A), with higher magnification. Scale bar: 1 µm. (C) Confocal x/z-scan of a membrane clad compartments with a flat bottom (left) vs a more tapered compartment (right). The membrane was labeled with DiI to visualize the profile of the membrane compartments. The width of the compartments is about 10 µm at the upper region. Small variations in the corresponding shapes of the compartments occurred during the fabrication process, but still supported oscillations and gradient formation of the Min proteins. (D) Confocal images of FtsZ-mts in compartments with a flat bottom (left) and in a more tapered compartment. Scale bar: 5 µm.
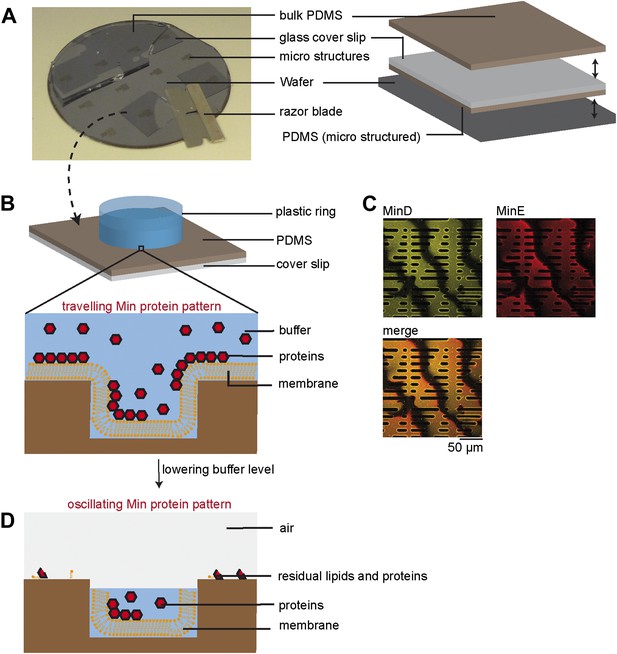
Experimental setup.
(A) A thick layer of PDMS was poured on top of a wafer with about 10 µm high resist microstructures. Glass cover slips were manually pressed in the liquid PDMS, such that an about 30 µm thin layer of PDMS remained below the glass surface and a thick layer of PDMS covered the glass surface on top. After curing the PDMS at 80°C the thick layer of PDMS on top was peeled off. Then the glass cover slip with the thin, micro structured layer of PDMS attached was peeled off together with the help of a razor blade. (B) After cleaning and plasma treading, a plastic ring was glued on top of the PDMS microstructures. Supported lipid membranes, which adapt to the topography of the underlying PDMS support, were produced using a vesicle fusion technique. Then proteins, ATP and/or GTP (depending on experiment) of defined concentration were added to a buffer volume of 200 µl on top of the supported lipid membranes. At this step of the protocol dynamic Min patterns formed on top of the membrane. These patterns did not oscillate but formed travelling waves. A Confocal image of Min waves on the upper level of the membrane support is shown in (C). Yellow: eGFP-MinD, red: MinE.Atto655. (D) Subsequently the buffer reservoir was manually reduced using a pipette, such that the buffer remained only in the micro fabricated chambers. Due to the confinement in small sample volumes with cell shaped geometry, the Min protein patterns oscillated within these chambers. The top of the chambers was open with a buffer/air interface. To limit evaporation of the buffer a lid was placed on top of the plastic ring. The upper level of the PDMS supports (outside of the compartments) was dry and residual biomolecules, such as remaining lipids and Min proteins were immobile, as determined by FRAP experiments.
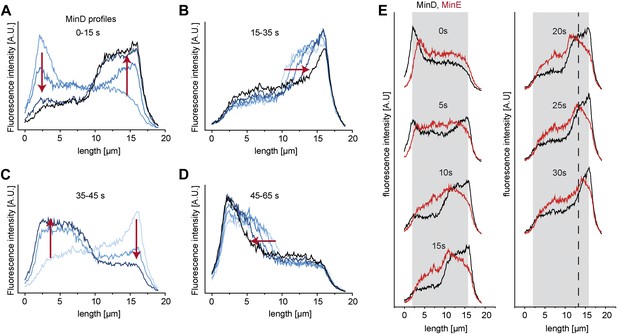
Protein gradients in vitro are established by dynamic redistribution of proteins.
(A–D) Concentration profiles of MinD along the length axis of a cell-shaped container were measured in intervals of 5 s. The whole oscillation cycle takes about 1 min. (A) Initially MinD forms a cap at the left pole of the compartment (light blue). Then the concentration of the MinD at the left pol decreases whereas it increases at the opposite pole. (B) The trail of the MinD zone at the right pol moves towards the right pole. (C) The concentration of MinD at the right pol decreases whereas it increases at the left pole. (D) The trail of the MinD cap at the left pole moves towards the left pole. (E) MinD (black) and MinE (red) concentration profiles during an oscillation from the left to the right pole. Dotted line: reference line to better visualize the movement of the MinE peak and MinD tail to the right pole.
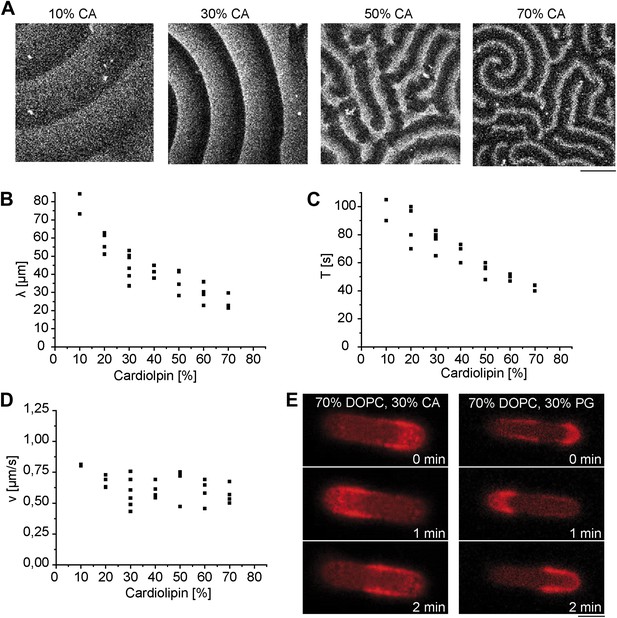
Lipid composition dependent formation of protein gradients.
(A) Pattern of MinD and MinE (labeled with Atto655) on supported lipid membranes with different amounts of cardiolipin. Scale bar: 50 µm. (B) Dependence of the Min pattern wavelength on cardiolipin concentration. (C) Dependence of temporal period on cardiolipin concentration. (D) Dependence of wave velocity on cardiolipin concentration. (E) Pole-to-pole oscillations emerge both in DOPC-membranes supplemented with cardiolipin and in DOPC-membranes supplemented with PG instead. Scale bar: 5 µm, DOPC: 1, 2-dioleoyl-sn-glycero-3-phosphocholine, CA: E. coli cardiolipin, PG: E. coli L-α-phosphatidylglycerol (Avanti polar lipids, Alabaster, AL).
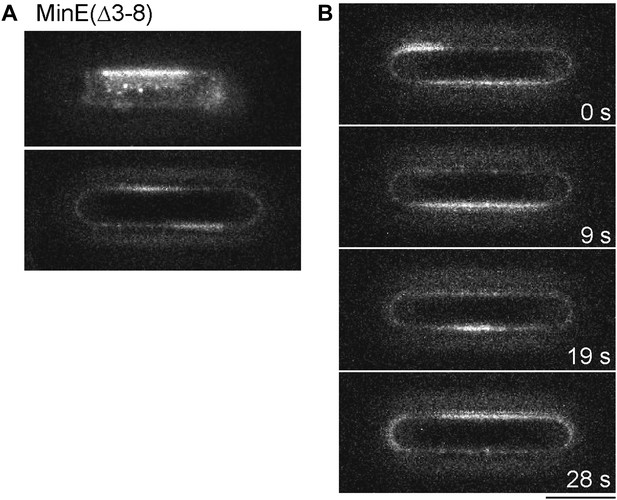
Deletion of the membrane targeting sequence of MinE affects pattern formation in micro compartments.
1 µM MinD and 1 µM MinE(Δ3–8) (5% labeled with Alexa Fluor 647) were reconstituted in micro compartments. (A) Snap shots of Min protein pattern at the bottom of the compartment (upper panel) and at the walls of the compartments (lower panel focus level in the middle of the compartment). (B) Time-lapse series of Min pattern within a compartment. Scale bar: 10 µm.
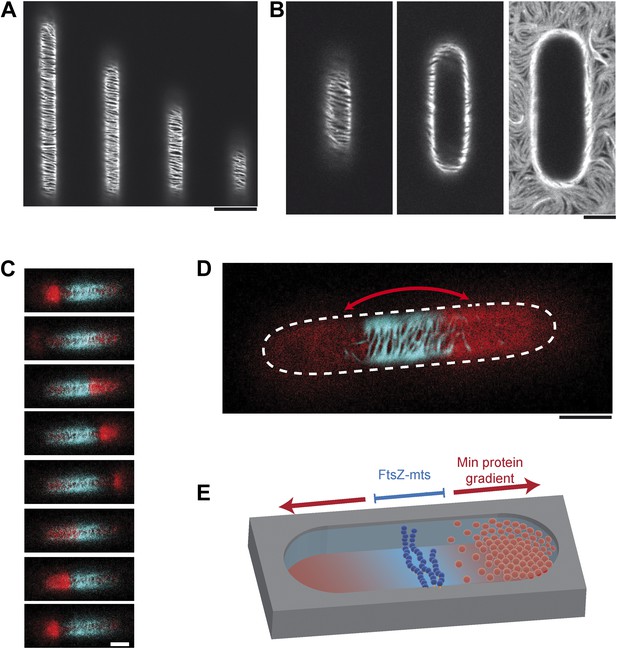
Reconstituted protein gradients coordinate localization of downstream targets in cell-shaped compartments.
(A) Confocal image of the tubulin homolog FtsZ-mts on the bottom of membrane-clad compartments. FtsZ-mts assembles into bundles which are aligned perpendicular to the length axis of the compartments. Scale bar: 10 µm. (B) Images with the focal plane at the bottom, middle and top of the compartment, respectively. The buffer was not yet reduced to a level below the upper level of the chip. Therefore membrane and FtsZ-mts network are still intact on the upper level of the chip. Scale bar: 5 µm. (C and D) When the gradient forming Min system (red: MinE.Atto655) and FtsZ-mts (blue) are reconstituted together, the Min gradient coordinates the localization of FtsZ-mts to the middle of the compartments. Scale bar: 5 µm. (C) Time between frames: 20 s. Negative controls in Figure 2—figure supplement 1 confirm that MinD and MinE are required to form dynamic patterns and that MinC is needed in addition to deplete FtsZ-mts from the membrane. (E) Schematic image of reconstituted FtsZ-mts positioning.
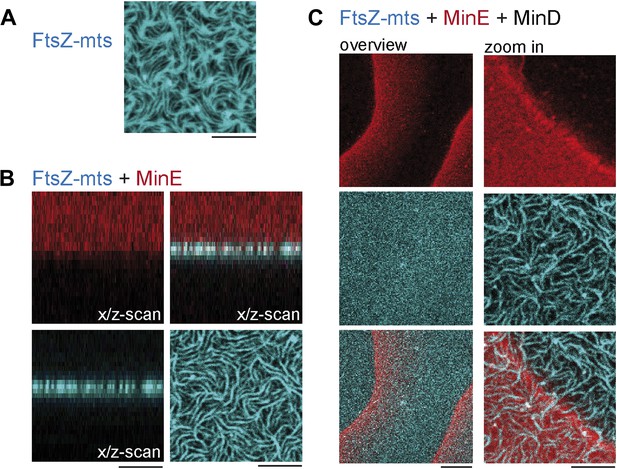
MinD MinE and ATP are required to form dynamic patterns and MinC is needed to deplete FtsZ from the membrane.
(A) Flat supported lipid membrane with reconstituted FtsZ-mts. Scale: 5 µm. (B) FtsZ-mts and MinE. Scale: 5 µm. (C) FtsZ-mts, MinE and MinD scale (overview): 20 µm, scale (zoom in): 5 µm.
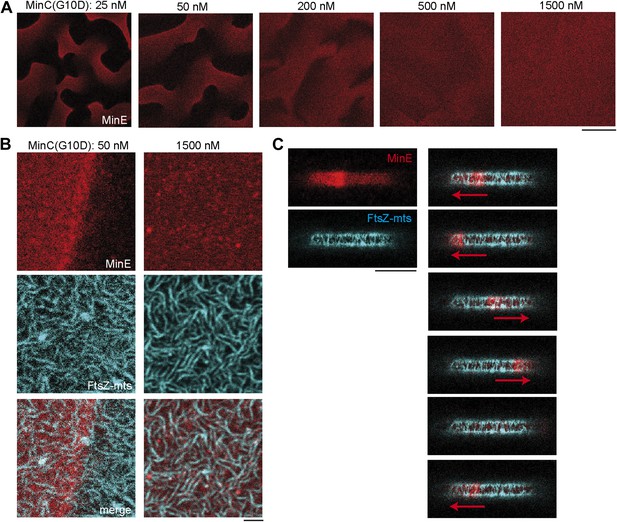
MinC-G10D does not inhibit polymerization of FtsZ-mts.
(A–C) FtsZ-mts (blue), MinD, MinE (5% labeled with Atto655: red) were reconstituted with the MinC-G10D mutant. Concentrations of MinD, MinE and FtsZ-mts are constant for all images. (A) Confocal images of Min protein pattern (MinE.Atto655: red) on flat supported membranes at different concentrations of MinC-G10D. High concentrations of MinC-G10D disturb Min protein patterns. The contrast between images is not comparable and is increased for higher concentrations of MinC-G10D, because the MinE intensity at the membrane decreased with higher concentrations of MinC-G10D. Scale bar: 50 µm. (B) Confocal images of the Min system (MinE.Atto655:red) and FtsZ-mts (blue) on flat membranes at different concentrations of MinC-G10D demonstrate that MinC-G10D is inefficient in disturbing FtsZ-mts networks. Scale bar: 2 µm. (C, Figure 3—figure supplement 1) In cell-shaped micro compartments with MinD, MinE, FtsZ-mts and 50 nM MinC-G10D the Min system (MinE.Atto655: red) oscillates from pole-to-pole. However MinC-G10D does not inhibit polymerization of FtsZ-mts (blue) at the compartment poles. Time between frames: 90 s, scale bar: 10 µm.
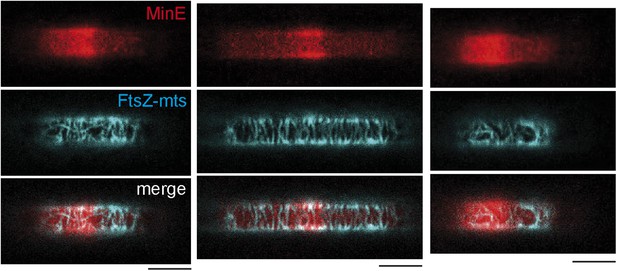
MinC-G10D does not inhibit polymerization of FtsZ-mts at compartment poles.
Confocal images of three compartments with FtsZ-mts (blue), MinD, MinE (5% labeled with Atto655: red) and 50 nM MinC-G10D in microcompartments. The FtsZ-mts bundels are not inhibited at compartment poles. Scale bar: 5 µm.
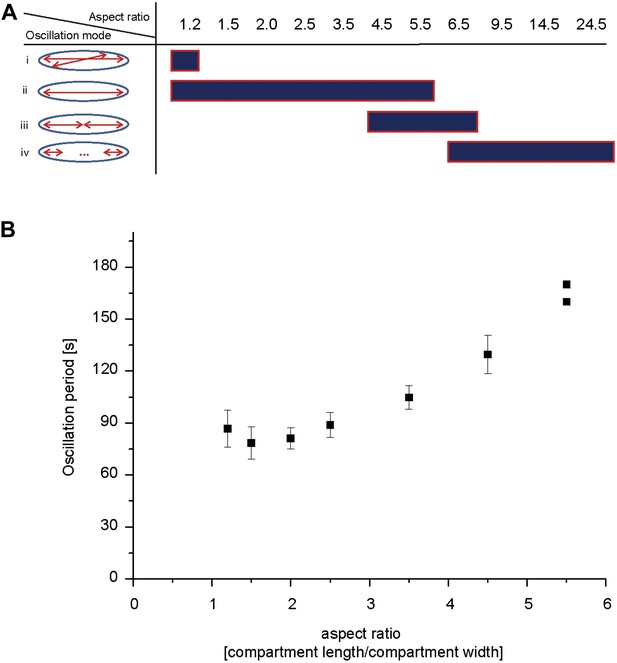
Compartment length dependent protein oscillations.
(A) Experiments such as in Video 1 were analyzed for the oscillation mode in the corresponding compartments and categorized as (i) oscillations with aberration from the long axis of the compartment, (ii) pole-to-pole oscillations, (iii) double oscillations, (iv) higher oscillation mode (at least triple oscillations). (B) Oscillation period of pole to pole oscillations in dependence of compartment length. Simultaneous time-lapse acquisition of Min protein patterns in multiple compartments allowed highly comparable experimental conditions. In total, 27 to 29 compartments of seven different aspect ratios (1.2, 1.5, 2.0, 2.5, 3.5, 4.5, 5.5) on the same time lapse movie were analyzed. Out of 27 compartments with an aspect ratio of 1.2, one compartment harboured oscillation that deviated from the length axis. Out of 28 compartments with an aspect ratio of 4.5 we observed 16 compartments, and out of 29 compartments with an aspect ratio of 5.5 we observed 27 compartments with double oscillations or oscillations which switched between pole-to-pole oscillations and double oscillations. All remaining compartments harboured pole-to-pole oscillations for which the mean oscillation period and standard deviation was calculated. For an aspect ratio of 5.5 the two data points are directly plotted, due to the small sample size. Figure 4—figure supplement 1 demonstrates that the oscillation period increases over time.
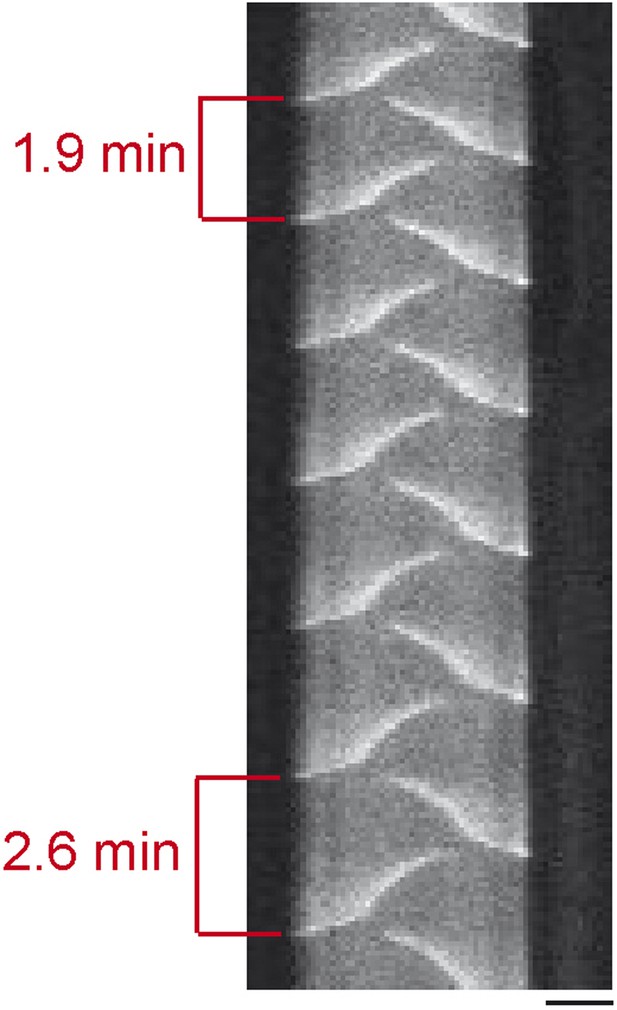
Kymograph of MinE oscillations.
Over time the Min protein oscillations become slower. This effect is most likely due to a decrease in the buffer level originating from the evaporation of buffer. 1 µM MinE (5% labeled with Atto.655), 1 µM MinD. Scale bar: 10 µm.
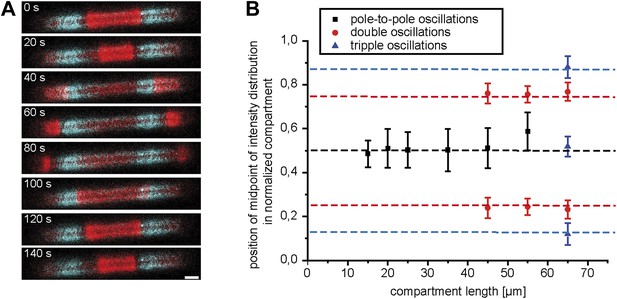
Compartment length affects FtsZ localization.
(A) In compartments with double oscillations of the Min system (red: MinE.Atto655), FtsZ-mts (blue) localizes to two regions along the cell length. Scale bar: 5 µm. (B) Localization of FtsZ-mts along the length axis of an artificial cell for pole-to-pole oscillations (black), double oscillations (red) and triple oscillations (blue) in comparison to expected values (dotted lines), considering the symmetries of oscillation modes of Min protein along the long axis of a compartment. For pole-to-pole oscillations in 55 µm long compartments: n = 3 (For 55 µm the majority of structures revealed double oscillations and only in a minor fraction of these long structured occured pole-to-pole oscillations). For all other values, n >9.
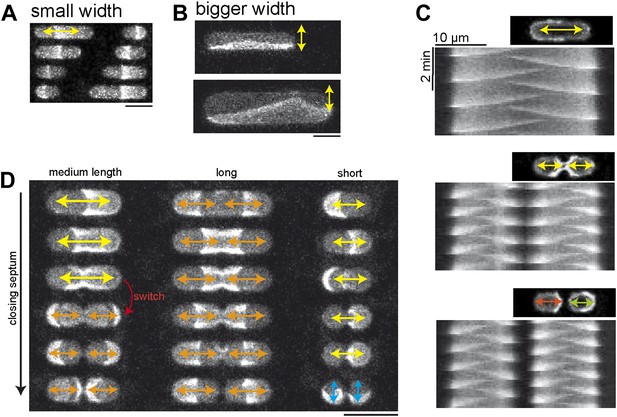
Compartment width affects gradient formation.
(A) Pole to pole oscillations occur is narrow compartments. Scale bar: 20 µm. (B) In wider compartments the proteins oscillate parallel to the length axis of the compartment. Scale bar: 20 µm. In compartments with an even larger width more complex patterns occur as depicted in Figure 6—figure supplement 1. (C) Confocal images and kymographs of MinE pattern in compartments with different degrees of ‘constriction’ depict pole-to-pole oscillation in compartments with a constant width (upper panel). In compartments with a narrow width in the middle of the compartment double ascillations occur. In separated compartments independent pole-to-pole oscillations are observed. (D) Oscillation modes in compartments with different lengths and stepwise narrowing width at the middle of the compartment. Left: Compartments without constriction or only slight decrease of the width in the middle harbor pole-to-pole oscillations (asymmetric oscillations, yellow arrows). Compartments with a narrow width in the middle harbor double oscillations (symmetric oscillations, orange arrows). A series of time-lapse images as well as kymographs of the third and fourth compartment, in which the transition from pole-to-pole to double oscillation occurs are shown in Figure 6—figure supplement 2. Video 5 shows the dynamic pattern of the third and fourth compartment in this image. Middle: In longer compartments double-oscillations occur due to the increased length, even if the width along the length of the compartment is constant. Right: In very short cells the constriction of the compartments results in two connected compartments, with length axis perpendicular to the length axis in non-constricting compartments. Thus the Min proteins oscillate along the new length axis, which is perpendicular to the oscillation axis in non-constricting short compartments (blue arrows). 1 µM MinE (doped with 10% MinE.Atto655), 1 µM MinD, Scale bar: 20 µm Figure 6—figure supplement 3 demonstrates that oscillations happen between each ‘constricting septum’ if the compartment harbors two septa.
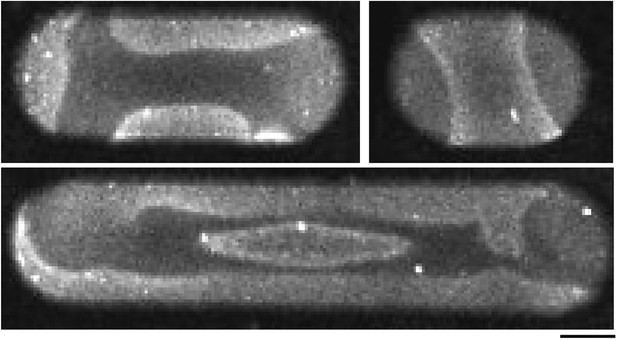
Min proteins self-assemble into complex pattern in large compartments.
Confocal time-lapse images of MinD and MinE (doped with 10% MinE.Atto655) in large compartments with a width of 50 µm. Scale bar: 20 µm.
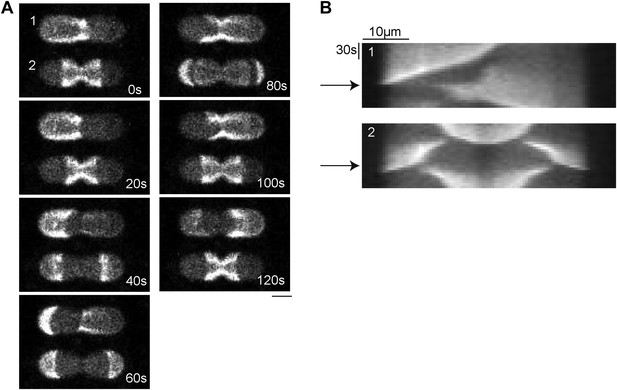
Min proteins patterns switch from asymmetric pole-to-pole oscillations to symmetric double oscillation when the neck in the middle of a compartment decreases.
(A) Confocal time-lapse images of MinD and MinE (doped with 10% MinE.Atto655) in compartments with decresing width of the neck from compartment 1 to 2. The depicted compartments at time step ‘80 s’ correspond to the third and fourth compartment in the left column of Figure 6D. Scale bar: 10 µm. (B) Kymographs of the compartments shown in (A). The arrows indicate the time (80 s) which corresponds to the frame in Figure 6D.
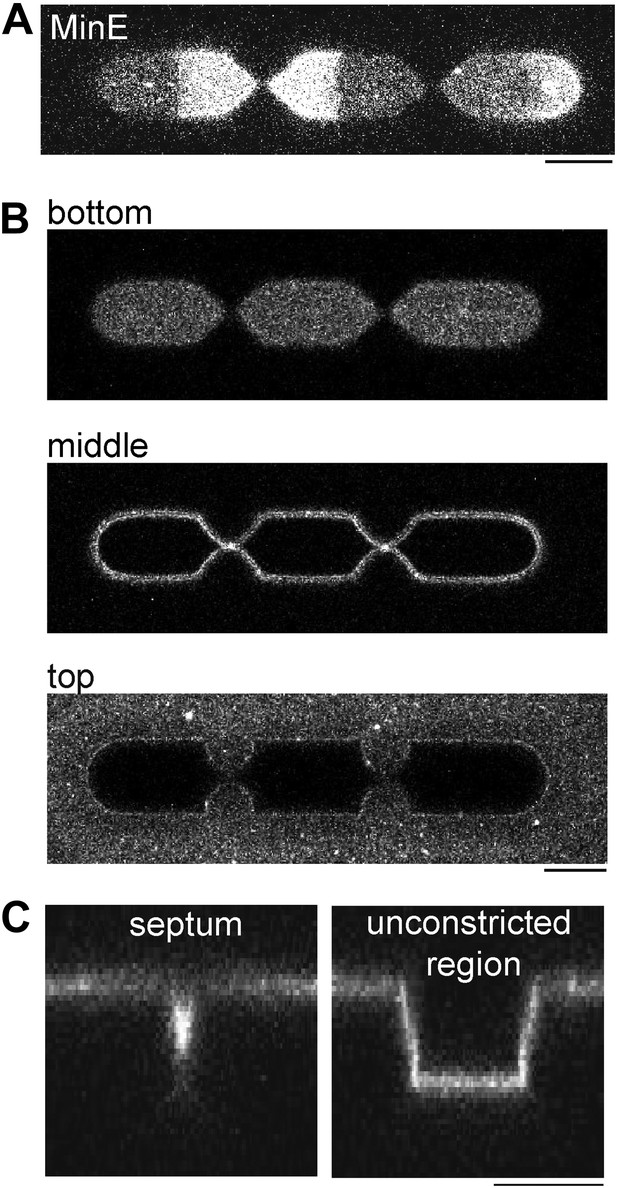
Min oscillations in compartments with multiple septa.
(A) Confocal image of MinD/E pattern in a compartment with two septa. The figure demonstrates that Min oscillations occur between each ‘constricting septum’. 5% MinE is labeled with Atto655. (B) The lipid membrane was labeled with DiI to visualize the surrounding of the compartment. The confocal images were taken with the focus level on the bottom, in the middle (5 µm above the bottom) and the upper level of membrane clad compartments (10 µm above the bottom of the compartment. (C) x/z-scans depict the profile of the compartment at the septum and at an unconstricted region. (A–C) Scale bar: 10 µm.
Videos
Reconstituted Min protein oscillations in microcompartments of different length.
This video shows a confocal time lapse movie of reconstituted MinD/MinE pattern in cell-shaped micro-compartments. 10% MinE are labeled with Alexa 488. The movie follows the dynamics of Min protein pattern for 17 min.
Min protein pattern in short compartments.
This movie shows confocal time-lapse movies of a MinD and MinE patterns (doped with 10% MinE.Alexa488) in 12 µm long and 10 µm wide compartments. Six time-lapse movies with compartments in which the Min patterns aberrate from a stable pole-to-pole axis are stitched together. The protein patterns are followed over a time of about 17 min.
Compartment width dependent orientation of Min oscillations.
This movie demonstrates how the oscillation axis switches depending on the width of the sample compartment. The confocal time-lapse movies of MinD and MinE patterns (doped with 10% MinE.Atto655) follows the dynamic oscillations over a time of 6 min.
Complex Min protein pattern in large compartments.
This movie shows a confocal time-lapse movie of a MinD and MinE patterns (doped with 10% MinE.Alexa488) in 50 µm wide compartments. The protein patterns are followed over a time of 6 min.
Min protein pattern in compartments with a small neck in the middle of the compartment.
This movie shows a confocal time-lapse movie of a MinD and MinE patterns (doped with 10% MinE.ATTO655) in compartments with a small neck in the middle. The width of the neck in the lower compartment is smaller as compared to the upper compartments and induces a switch from pole-to-pole oscillation to double oscillations. Time of the movie: 2 min.





