Reliable cell cycle commitment in budding yeast is ensured by signal integration
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record published
- Accepted Manuscript updated
- Accepted Manuscript published
- Accepted
- Received
Decision letter
-
James FerrellReviewing Editor; Stanford University, United States
eLife posts the editorial decision letter and author response on a selection of the published articles (subject to the approval of the authors). An edited version of the letter sent to the authors after peer review is shown, indicating the substantive concerns or comments; minor concerns are not usually shown. Reviewers have the opportunity to discuss the decision before the letter is sent (see review process). Similarly, the author response typically shows only responses to the major concerns raised by the reviewers.
Thank you for sending your work entitled “Reliable cell cycle commitment in budding yeast is ensured by signal integration” for consideration at eLife. Your article has been favorably evaluated by Richard Losick (Senior editor) and a member of the Board of Reviewing Editors (James Ferrell), and has been reviewed in depth by three peer reviewers.
The Reviewing editor and the reviewers discussed their comments before we reached this decision, and the Reviewing editor has assembled the following comments to help you prepare a revised submission.
The manuscript “Reliable cell cycle commitment in budding yeast is ensured by signal integration” by Liu et al. addresses the important problem of how yeast cells make the decision to commit to a new round of cell division at the Start transition. The authors reach the interesting conclusion that cells integrate Cln3 activity over time to make a reliable decision. This is in principle an important finding and the integration model could be very general.
We have two main criticisms that need to be addressed prior to publication. There are a number of minor criticisms as well, which would improve the rigor and understandability of the paper.
Major concerns:
1) One major concern is that the integration model is not compared to alternative models. I would suggest that the authors use statistical methods to prove that the integration model provides a significantly better fit of the experimental data than alternative models (for example instantaneous Cln3 concentration or instantaneous total Cln3 in the whole cell). Notice that equation number 4 in the Supplementary file 1 actually suggests that the total amount of Cln3 in a cell should be a good estimator of when cells divide. The authors need to provide stronger arguments to distinguish among models. This should be readily doable, as they have already acquired the necessary data.
2) An important assumption of the integrator model is that Whi5 has a 'memory', namely that its activity depends not only on the instantaneous Cln3 but also on previous events. This means that its dephosphorylation rate is slow. In the supplementary materials, the authors cite published work from Charvin et al. (2010) that they say indicates that the half-time for Cln3 dephosphorylation is longer than 30 min. However this is not exactly what Charvin et al. said or what Charvin et al.'s data show.
What Charvin et al. showed (their Figure 4E) is that following a 15 min pulse of CLN2 expression in the presence of Clb-inhibitory levels of Sic1-4A expression, Whi5 exits the nucleus and then begins to reenter it. Re-entry begins very soon after the inducer of CLN2 expression is washed away and is half-maximal by 35 min. Some of this time interval-Charvin et al. guess about 5-10 min-is probably due to the kinetics of CLN2 degradation. Presumably the remaining ∼25-30 min is partly attributable to the kinetics of Whi5 dephosphorylation, and partly to the kinetics of Whi5 nuclear transport. So based on these experiments, one might conclude that Whi5 dephosphorylation could have a half-time of up to 30 min, not that it has a half-time of longer than 30 min.
There are two ways the authors could deal with this issue. They could directly measure for themselves how fast Whi5 returns to the nucleus after inhibition of CDK1. One way would be to use an analog-sensitive CDK1 mutant, which would obviate the need to guess how long CLN2 remains above the level required for Whi5 phosphorylation after the induction of CLN2 is terminated. While this might be the ideal solution, we appreciate that there is already a lot of data in the paper, and we would be satisfied if the authors would just accurately state Charvin et al.'s findings in the main text and acknowledge that slow Whi5 dephosphorylation kinetics remains a key incompletely-tested assumption of the authors' model.
https://doi.org/10.7554/eLife.03977.031Author response
1) One major concern is that the integration model is not compared to alternative models. I would suggest that the authors use statistical methods to prove that the integration model provides a significantly better fit of the experimental data than alternative models (for example instantaneous Cln3 concentration or instantaneous total Cln3 in the whole cell). Notice that equation number 4 in the Supplementary file 1 actually suggests that the total amount of Cln3 in a cell should be a good estimator of when cells divide. The authors need to provide stronger arguments to distinguish among models. This should be readily doable, as they have already acquired the necessary data.
We acknowledge the referees’ suggestion.
When deducting equation number 4 in the Supplementary file 1, we assumed Cln3 concentration is constant during G1 phase. G1 length is inversely proportional to average Cln3 concentration, which equals to instantaneous Cln3 concentration under this assumption. We clarified the assumption in Supplementary file 1.
Start is triggered when phosphorylated Whi5, Whi5p, reaches the threshold Whi5p(G1)=Whi5tot-Whi5c. When considering the fluctuation of Cln3 concentration with time,
G1 length cannot be estimated by the instantaneous Cln3 concentration at one time point but by the Cln3 integration through the whole G1 phase. We added an additional section in Supplementary file 1A to discuss how G1 length is determined when relaxing the assumption of constant Cln3 concentration.
We tested the Instantaneous Model with the Cln3 profiles measured in experiment, which are more meaningful than stochastic simulations. In our experimental setup, Cln3 level during G1 in one cell fluctuates with about 20% CV (see Author response image 1A). If the Instantaneous Model were true, cells should pass Start at or near the peak of Cln3 profile. However, we found that in near 80% cells, the timing of Cln3 peak is different from the timing of Start (see Author response image 1B). Even when we relax the timing requirement of the two events to be within 10 min, there are still over 50% cells that failed the test. The result is even more significant when focusing on G1 lengths longer than 30 min (see Author response image 1C). It is thus very unlikely that Start is triggered by the instantaneous Cln3 concentration. We added this result as Figure 2–figure supplement 2 and a corresponding explanation in the main text. A major difference between the Instantaneous Model and the Integration Model is the memory length. The two models are equivalent in the limit of zero memory. We estimated the memory length to be over 10 min (Figure 2–figure supplement 3), comparable to the G1 length in daughter cells (See the next response).
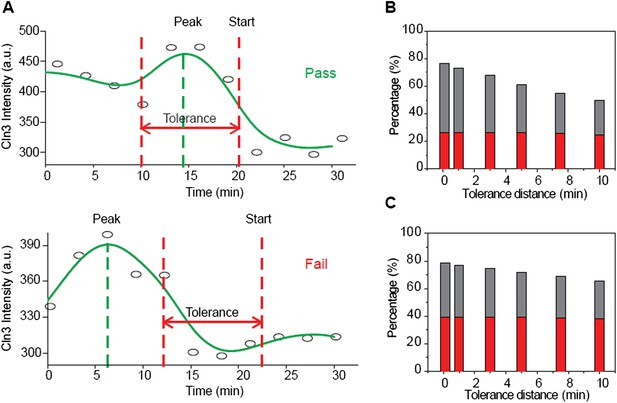
The Instantaneous Model fails with the Cln3 profiles measured in experiment. (A) Schematic plot of the test. The test is considered a pass if the timing of Start is near the timing of Cln3 peak by the specified tolerance value. The Cln3 profiles are from the real data. Open circles denote the raw data; solid lines are the smoothing splines. (B-C) Test failure percentage. Grey bars indicate the failure percentage and red bars indicate the percentage of cells whose Cln3 peak value is more than 20% larger than Cln3 at Start, for all cells (B) and cells with G1 longer than 30 min (C).
2) An important assumption of the integrator model is that Whi5 has a 'memory', namely that its activity depends not only on the instantaneous Cln3 but also on previous events. This means that its dephosphorylation rate is slow. In the supplementary materials, the authors cite published work from Charvin et al. (2010) that they say indicates that the half-time for Cln3 dephosphorylation is longer than 30 min. However this is not exactly what Charvin et al. said or what Charvin et al.'s data show.
What Charvin et al. showed (their Figure 4E) is that following a 15 min pulse of CLN2 expression in the presence of Clb-inhibitory levels of Sic1-4A expression, Whi5 exits the nucleus and then begins to reenter it. Re-entry begins very soon after the inducer of CLN2 expression is washed away and is half-maximal by 35 min. Some of this time interval-Charvin et al. guess about 5-10 min-is probably due to the kinetics of CLN2 degradation. Presumably the remaining ∼25-30 min is partly attributable to the kinetics of Whi5 dephosphorylation, and partly to the kinetics of Whi5 nuclear transport. So based on these experiments, one might conclude that Whi5 dephosphorylation could have a half-time of up to 30 min, not that it has a half-time of longer than 30 min.
There are two ways the authors could deal with this issue. They could directly measure for themselves how fast Whi5 returns to the nucleus after inhibition of CDK1. One way would be to use an analog-sensitive CDK1 mutant, which would obviate the need to guess how long CLN2 remains above the level required for Whi5 phosphorylation after the induction of CLN2 is terminated. While this might be the ideal solution, we appreciate that there is already a lot of data in the paper, and we would be satisfied if the authors would just accurately state Charvin et al.'s findings in the main text and acknowledge that slow Whi5 dephosphorylation kinetics remains a key incompletely-tested assumption of the authors' model.
We acknowledge the referees’ correction for our interpretation of Charvin et al.'s data. To better understand the integration mechanism, we measured Whi5 dephosphorylation kinetics as the referees suggested. As most Whi5 is dephosphorylated and resides in the nucleus during G1 phase, we could not directly measure Whi5 dephosphorylation in G1. Thus we measured Whi5 nuclear entry right after G1 by inhibiting Cdk1 activity with a strain bearing a cdc28-as1 allele. The average half time of Whi5 nuclear entry, which corresponds to the memory length in the mathematical model, is 13.7 min in mother cells and 10.6 min in daughter cells, respectively. Note that the average G1 length (from cytokinesis to Start) in daughter cells is 13.6 min in SD medium, a memory length of about 10 min should be long enough for the cell to take the advantage of integration.
We added this result as Figure 2–figure supplement 3, and the corresponding explanation in the main text.
https://doi.org/10.7554/eLife.03977.032




