Competition between antagonistic complement factors for a single protein on N. meningitidis rules disease susceptibility
Figures
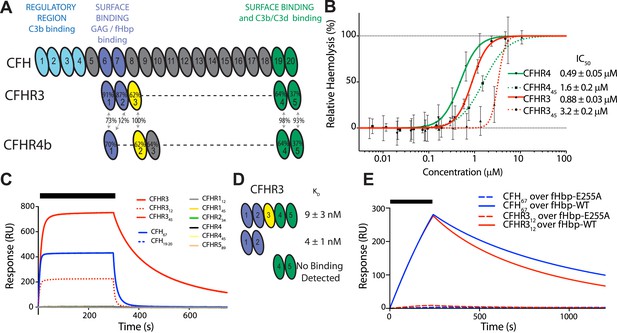
CFHR3 binds N. meningitidis fHbp at the same site as CFH and promotes complement activation.
(A) Complement control protein (CCP) domains of CFH, CFHR3, and CFHR4 are shown with the sequence identity to CFH and between the CFHRs indicated. Key functional regions of CFH are noted. (B) CFH-dependent alternative pathway (AP) haemolytic assay (Goicoechea de Jorge et al., 2013). Using a CFH dose that reduced lysis of guinea pig erythrocytes to 50%, the addition of increasing concentrations of either full-length or C-terminal fragments of CFHR3 or CFHR4 resulted in a dose-dependent increase in lysis. The two N-terminal CCP domains from CFHR3 had no effect (not shown). (C) Surface plasmon resonance (SPR) was used to investigate interactions of CFHRs with fHbp. Full-length and fragments of CFH and CFHRs were injected (black bar) over surface bound fHbp3.28. Only CFHR312, full-length CFHR3, or CFH67 bind to fHbp. (D) Dissociation constants (KD) measured by SPR (CFHR312) or microscale thermophoresis (CFHR3) indicated the fHbp binding site is within the two N-terminal domains of CFHR3. (E) SPR with 200 nM protein flowing over V3.28 fHbp demonstrates that the single point mutation E255A, which ablates CFH binding, also ablates CFHR12 binding.
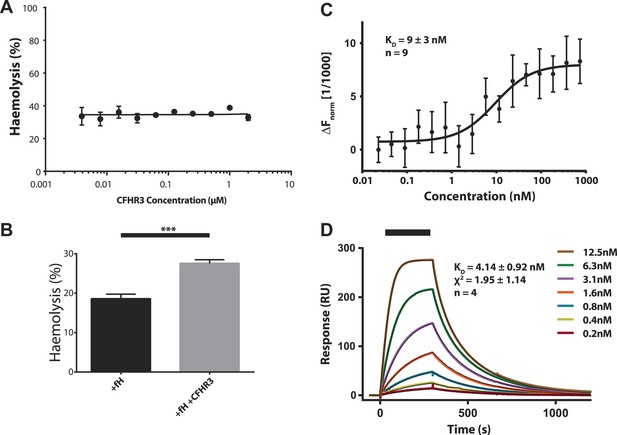
Increased haemolysis on addition of CFHR3 is dependent on presence of CFH and interaction of CFHR3 full length and N-terminal fragment with V3.28 fHbp.
(A) Haemolysis of guinea pig erythrocytes by 5% CFH depleted serum (CompTech, TX, USA) was measured upon addition of a range of CFHR3 concentrations (4 nM–2 μM) in Mg-EGTA to ensure activation by the alternative pathway (AP) alone. Haemolysis is presented as a percentage of maximal lysis by H2O. No significant differences in haemolysis were observed under any of the conditions. (B) Supplementing the CFH deficient serum (CompTech, TX, USA) with 100 nM CFH reduced lysis to ∼20%. In this CFH-supplemented experiment addition of 2 mM CFHR3 led to a significant (p = 0.003, two-tailed, unpaired t test) increase in haemolysis as seen with the naturally CFH sufficient serum in Figure 1. (C) Full-length CFHR3 interacts strongly with the carboxymethyldextran matrix used to immobilise proteins in SPR, therefore, microscale thermophoeresis (Seidel et al., 2013), a solution technique, was used to measure the affinity for V3.28 fHbp. The data plotted are mean ± SE for nine repeats, which were simultaneously fit to derive the KD and error estimate shown. (D) SPR was used to measure the KD of the interaction with V3.28 fHbp (see ‘Materials and methods’). Data from a single experiment are shown whilst the KD and fit statistics are mean ± SD for four independent repeats.
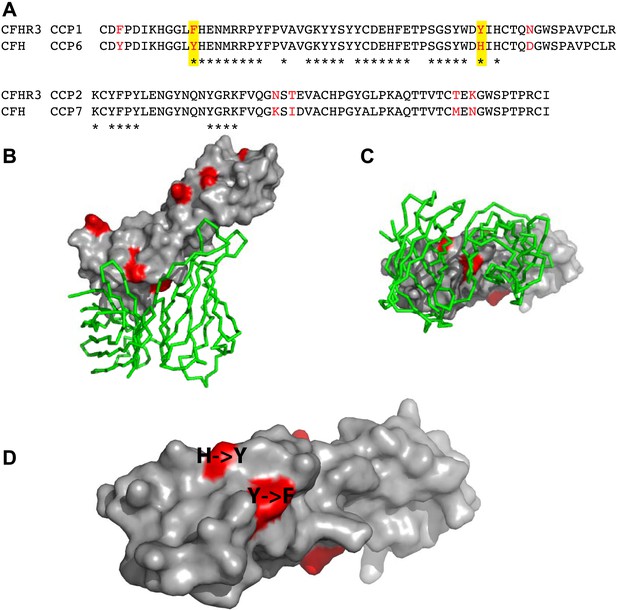
Amino acid differences between CFHR3 and CFH in the fHbp binding site.
(A) Sequence alignment between CFHR312 and CFH67. Differences are highlighted in red text, the two differences within the fHbp binding site are highlighted in yellow and residues contacting fHbp in the CFH-fHbpV3 structure (PDB ID 4AYM [Johnson et al., 2012]), chains A and D are indicated by an asterisk below the CFH sequence. (B) CFH67 is shown as a grey surface with residues that differ in CFHR3 highlighted in red; fHbp is shown as a green ribbon. (C) Rotated 90° around x compared to the view in B. (D) Looking down onto the fHbp binding site on CFH the two, conservative, sequence differences between the CFH67 and CFHR312 found in the binding site are indicated in red on the grey surface.
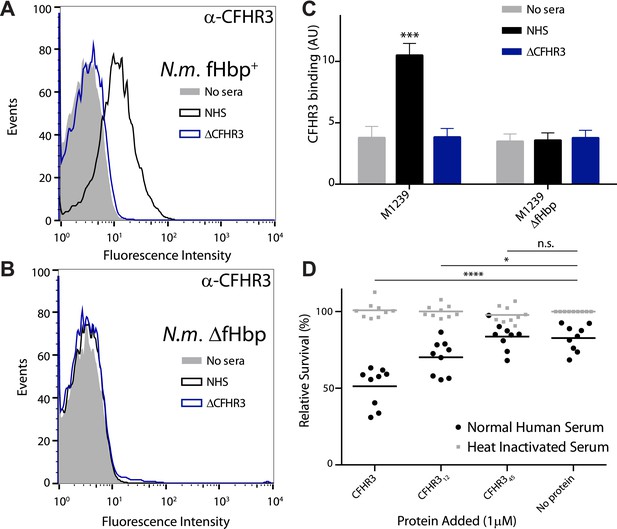
N. meningitidis binds CFHR3 on its surface in an fHbp-dependent manner promoting complement-mediated lysis.
(A–C) Flow cytometry of N. meningitidis demonstrates that CFHR3 binds to fHbp on the bacterial surface. (D) Sensitivity of N. meningitidis strain M1239 to complement-mediated cell lysis after pre-incubation with 1 µM CFHR3, CFHR312, CFHR345, or no protein. Faded symbols are bacteria incubated in identical conditions except using heat-inactivated NHS instead of NHS. Data presented as percentage survival relative to bacteria incubated in heat-inactivated NHS without additional CFHR3. Line shows the mean of three independent biological experiments, each with three technical replicates; individual results indicated. Significance was calculated using a two-tailed unpaired t test comparing with values obtained with no additional protein: ****p < 0.0001; *p < 0.05; ns p > 0.05.
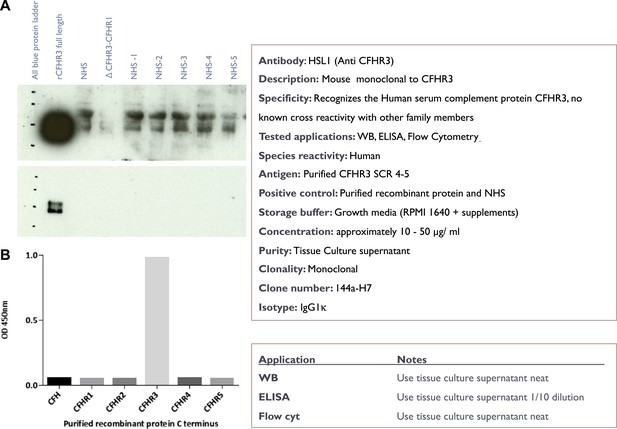
Characterisation of the novel anti-CFHR3 antibody HSL1.
(A) Western blotting of recombinant CFHR3 and sera from individuals sufficient or deficient in CFHR3 demonstrates that HSL1 is specific for CFHR3. (B) For screening of mAbs, ELISA plates (F96 maxisorp, Nunc) were coated with C-terminal pair of CCP domains of CFH and the CFHRs (5 µg/ml, 50 µl per well) overnight at room temperature before blocking with 2% skimmed milk in PBS 0.05% Tween 20. Plates were incubated with neat hybridoma culture supernatant and binding was detected with goat anti-mouse HRP (1 in 5000, Dako) followed by ELISA substrate and stop solution (Roche).
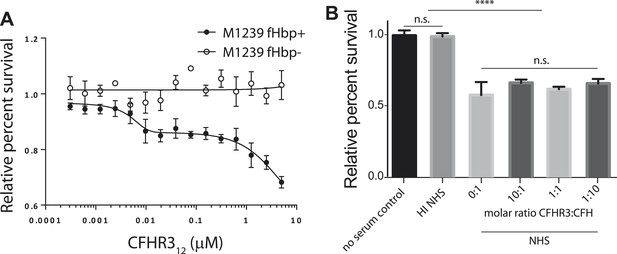
The effect of CFHR312 on bacterial survival is dose and fHbp dependent.
(A) Survival of N. meningitidis M1239 (filled circles) and M1239Δfhbp (unfilled circles) in the presence of NHS following pre-incubation in different concentrations of CFHR312 (indicated). Data presented as percent survival relative to bacteria incubated in the absence of serum and CFHR312. Error bars are ±SEM of three independent experiments. (B) Serum survival of E. coli DH5α (which does not express a CFH-binding surface protein) is not altered by pre-incubation with CFHR3. Data presented as percent survival relative to bacteria incubated in the absence of serum and CFHR3. Error bars are ±SEM of three separate experiments repeated four times. Significance was calculated using a standard one-way ANOVA with Tukey's multiple comparison test (n.s., not significant; ****p < 0.0001).
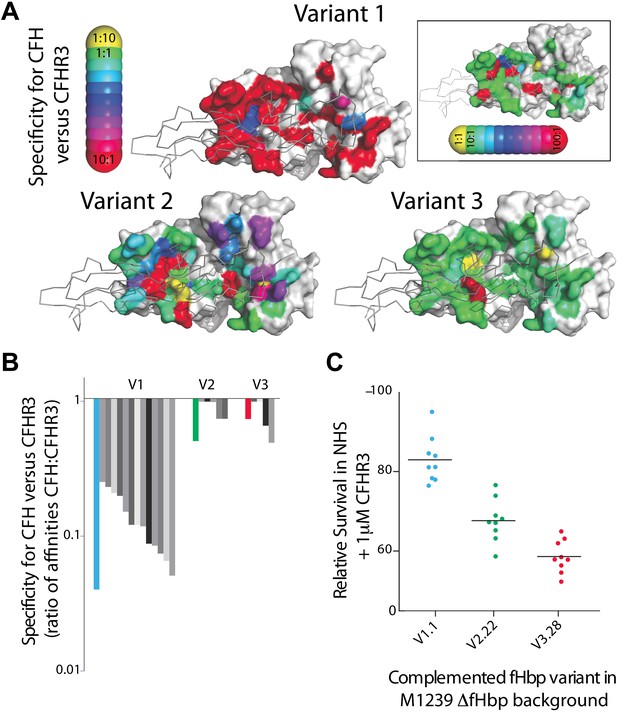
Key residues in fHbp that mediate binding to CFHR3.
(A) SPR was used to determine KDs for both CFH67 and CFHR312 binding to a panel of fHbps bearing alanine substitutions within the CFH67 binding site. ∼50 mutations were screened in each of three fHbps (V1.1, 2.21 and 3.28 [Johnson et al., 2012]). Mutations altered KDs for both CFH67 and CFHR312 but some mutations led to some specificity for binding one or the other protein (see full data in Extended Data File 1). (B) KDs were determined for a panel of fHbps from disease causing N. meningitidis. V1 fHbps have some ability to preferentially bind CFH67 vs CFHR312, whilst V2 and V3 fHbps were less able to distinguish the molecules (full data in Extended Data File 2). (C) AP-mediated bacterial killing of isogenic strains expressing different fHbps in the presence of CFHR3. Data are coloured as in panel (B) and bacterial survival in serum is increased in the strain expressing the fHbpV1.1 (that has the maximal specificity for CFH vs CFHR3).
-
Figure 4—source data 1
Excel spreadsheet with full details of KDs for CFH and CFHR312 derived from 1:1 Langmuir fits of SPR data for alanine scanning mutants and natural variant fHbp sequences.
- https://doi.org/10.7554/eLife.04008.010
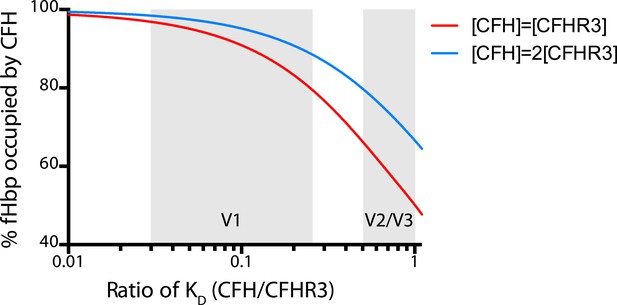
Modelling the occupancy of bacterial cell surface fHbp assuming different levels of specificity for CFH over CFHR3.
Each molecule of fHbp on the bacterial surface can bind with one molecule of CFHR3 or one molecule of CFH. Assuming that the number of molecules of either of these is significantly greater than the number of molecules of fHbp then the percentage of bacterial surface fHbp molecules binding CFH at equilibrium will be determined by: the relative amounts of both present in the individual; and the affinities of that fHbp sequence for CFH vs CFHR3 (essentially a model of competitive antagonism, see equation below). Natural fHbp sequences are relatively constant in their KD for CFH (KD ∼3 nM [Johnson et al., 2012] and this work), therefore we modelled the effect of the KD for CFHR3 varying between 3 nM and 300 nM against this constant KD for CFH under two conditions. In red we assume that both CFH and CFHR3 are present in serum at 1.5 μM, whilst in blue we model the effect if the concentration of CFH is twice that of CFHR3 (i.e., 3 μM vs 1.5 μM). This range of relative concentrations covers those generally reported for CFH and CFHR3 (except in the case of ΔCFHR3/1 individuals where the concentration of CFHR3 will be zero and 100% of the fHbp will bind CFH). The grey shaded areas indicate the regions of the x axis into which the families of natural variant fHbp sequences studied in Figure 3 fall. The equation used to generate these curves is:
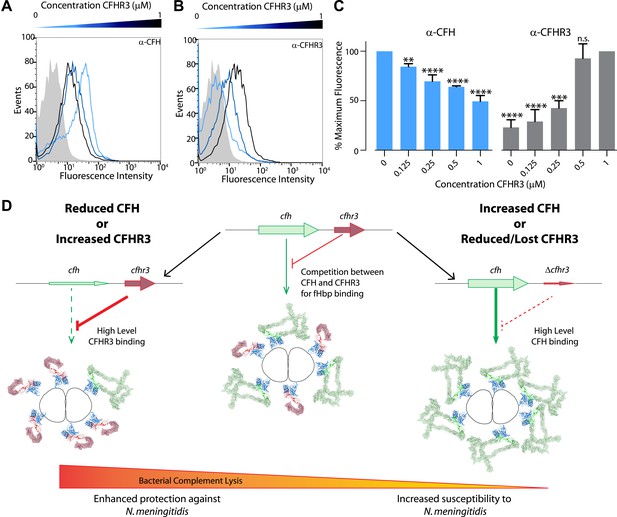
Susceptibility to N. meningitidis will be altered by the circulating levels of CFH and CFHR3 that compete for binding to fHbp.
(A–C) Flow cytometry demonstrates that addition of increasing amounts of CFHR3 (from 0 μM [light blue] to 1 μM [black]) to sera from a ΔCFHR3–1 individual leads to decreasing levels of CFH on the bacterial surface (A and C) and increasing levels of CFHR3 (B and C). (D) Model of how differing levels of CFHR3 and CFH in an individual combine to alter the amount of CFH hijacked to the bacterial surface, hence the level of complement-mediated bacterial killing and an individual's susceptibility to meningococcal disease.
Tables
Primers used to generate bacterial strains
| Primer name | Strain amplified | Ref | Primer Sequence (restriction site underlined) |
|---|---|---|---|
| pgcc4V1.1 F | H44/76 | (Jongerius et al., 2013) | CGGTTAATTAAGGAGTAATTTTTGTGAATCGAACTGCCTTCTGCT |
| pgcc4V1.1 R | H44/76 | (Jongerius et al., 2013) | CGGTTAATTAATTATTGCTTGGCGGC |
| pgcc4V1.14 F | NZ98/254 | CGGTTAATTAAGGAGTAATTTTTGTGAACCGAACTGCC | |
| pgcc4V1.14 R | NZ98/254 | CGGTTAATTAATTATTGCTTGGCGGCAAGAC | |
| pgcc4V2.22F | FAM18 | CGGTTAATTAAGGAGTAATTTTTGTGAACCGAACTGCCTTCTGCT | |
| pgcc4V2.22R | FAM18 | CGGTTAATTAACTACTGTTTGCCGGCGATGC | |
| pgcc4V3.28 F | M1239 | CGGTTAATTAAGGAGTAATTTTTGTGAATCGAACTGCCTTCTGCT | |
| pgcc4V3.28 R | M1239 | CGGTTAATTAACTACTGTTTGCCGGCGATGC |





