The extraembryonic serosa is a frontier epithelium providing the insect egg with a full-range innate immune response
Figures
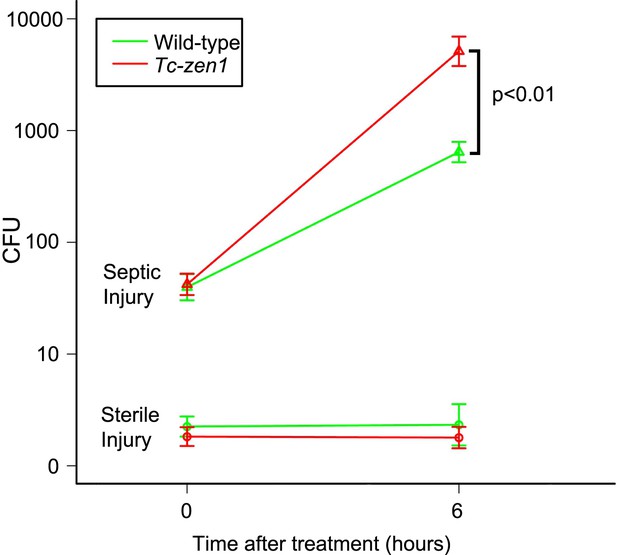
Counts of colony forming units (cfu's) after sterile and septic injury.
Green lines represent bacterial growth in wild-type eggs. Red lines represent bacterial growth in Tc-zen1 RNAi (serosa-less) eggs. Sterile injury did not introduce bacteria (lower lines: average of 2 cfu's found at t = 0 and an average of 5 cfu's found at t = 6). Septic injury introduced on average 53 bacteria into wild-type eggs and 49 into serosa-less eggs. These numbers increased to 747 ± 106 cfu's in wild-type eggs (green upper line) and to 7260 ± 1698 cfu's in serosa-less eggs (red upper line) at t = 6. This means that bacteria propagate twice as fast in serosa-less eggs (p < 0.01, as determined by a Pearson's chi-square test). Suspensions of 10 eggs were used per LB agar plate (see ‘Materials and methods’), and 10 plates were analyzed per treatment and time point, giving rise to the error bars presented in the graph (standard error).
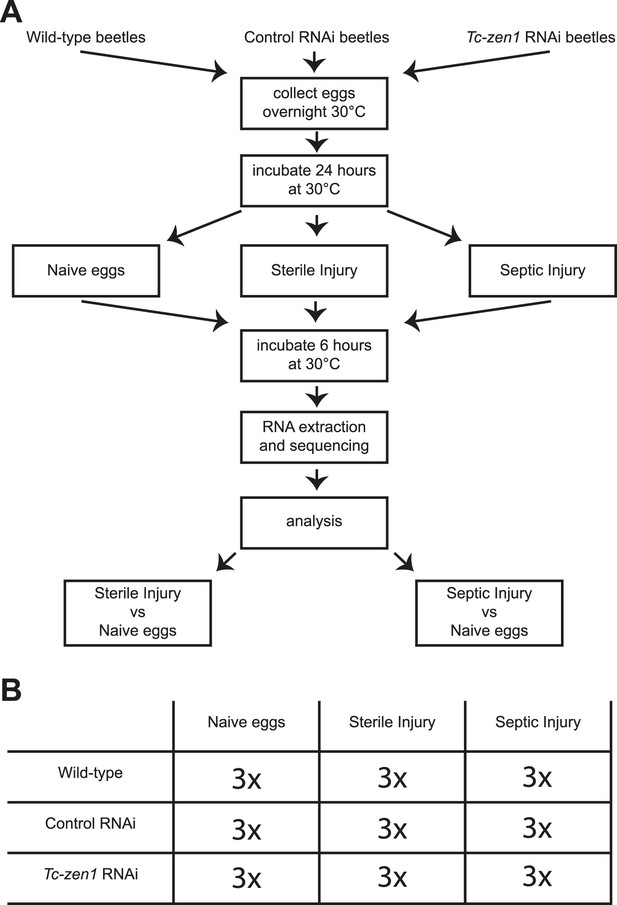
Experimental setup.
(A) We collected eggs from wild-type, control RNAi, and Tc-zen1 RNAi beetles overnight. These eggs were incubated for 24 hr at 30°C to ensure development of the serosa. Eggs are then maximally 40 hr old, while total developmental time is close to 85 hr at 30°C. Eggs were pricked with a sterile needle (sterile injury), pricked with a mix of E. coli and M. luteus (septic injury), or remained untreated (naive). They were incubated for another 6 hr at 30°C before total RNA was extracted for RNAseq. To analyze the immune response, the transcriptomes of sterilely injured eggs and of septically injured eggs were compared to naive eggs. This was done for wild-type, control, and Tc-zen1 RNAi eggs. (B) We collected three biological samples for each combination of egg-type (wild-type, control RNAi, or Tc-zen1 RNAi) and treatment (naive, sterile injury, or septic injury) giving a total of 27 biological samples.
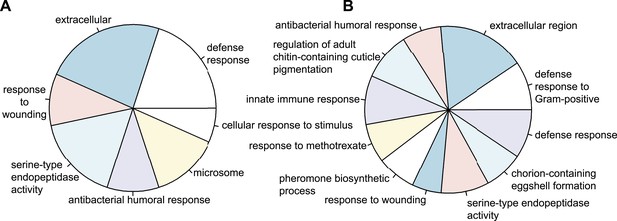
Types of genes that are differentially regulated.
(A) Significantly over-represented GO-terms among the genes induced in wild-type eggs after septic injury (p < 0.001). (B) Significantly over-represented GO-terms among the genes induced in control RNAi eggs after septic injury (p < 0.001). These categories indicate that the detected differential regulation does not result from artefacts induced by treatments (such as death or delayed development) and show that Tribolium eggs display an elaborate immune response.
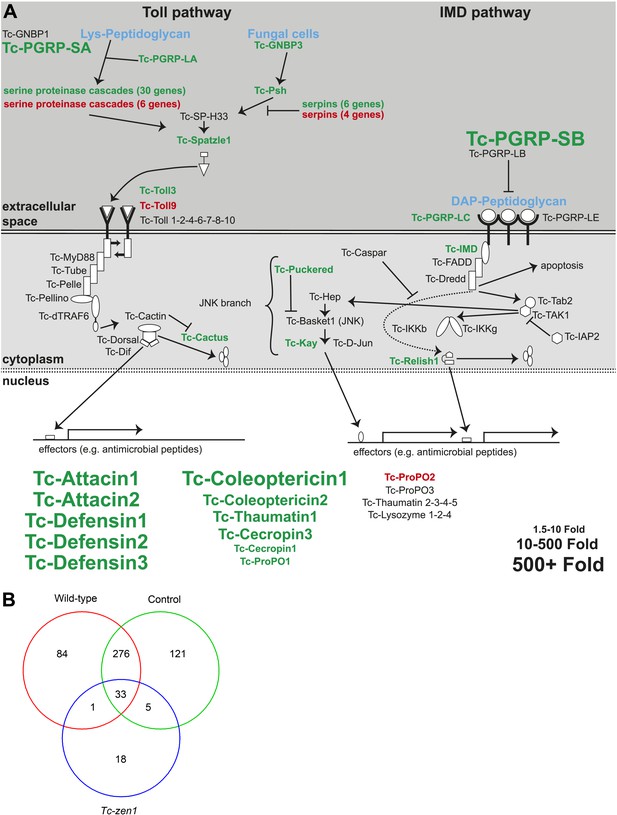
Immune-responsive genes in wild-type, control, and Tc-zen1 RNAi eggs.
(A) Schematic representation of the immune signaling pathways in Tribolium as described in Zou et al. (2007). Significantly induced genes after septic injury in wild-type or control RNAi eggs are indicated in green; significantly repressed genes after septic injury in wild-type or control RNAi eggs are indicated in red. Genes not differentially expressed are black. The size of the gene names represents the fold change (small = 1.5- to 10-fold, medium = 10- to 500-fold, large = 500 + fold expression). (B) Venn diagram showing the number of differentially expressed genes in septically injured eggs as compared to naive eggs (FDR < 0.01). In total, 538 genes are differentially expressed upon infection, of which 394 in wild-type eggs, 435 in control RNAi eggs, and only 57 in Tc-zen1 RNAi eggs. This means that Tribolium eggs display an extensive transcriptional response upon infection and that this response is largely abolished in eggs without a serosa.
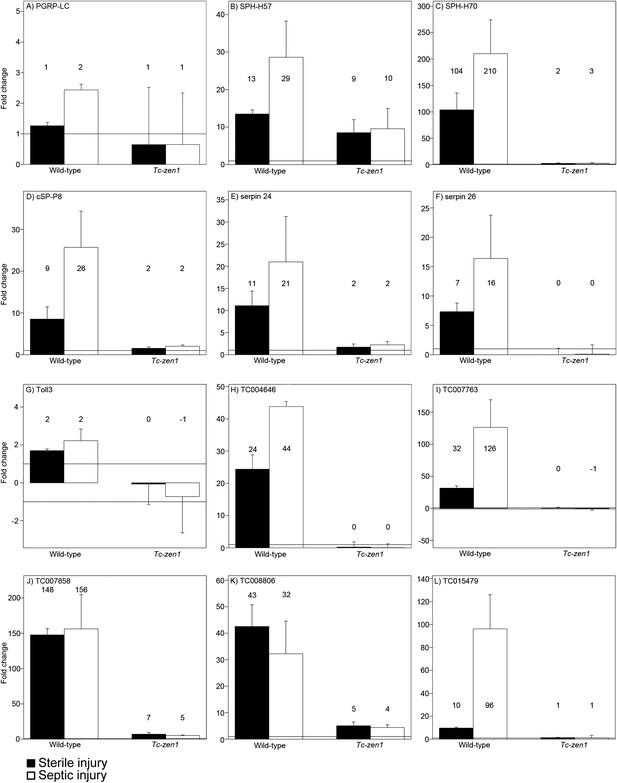
RT-qPCR verification of immune gene expression.
The expression levels of several immune genes was verified by RT-qPCR. Expression shown relative to the expression in naive eggs, the mean fold change of the biological replicates (based on two technical replicates) is plotted and error bars show the standard error. Black bars represent expression after sterile injury, white bars represent expression after septic injury. Expression levels measured by RT-qPCR show very similar results as the expression levels measured by RNAseq (See Supplementary file 2). (A) PGRP-LC, (B) SPH-H57, (C) SPH-H70, (D) cSP-P8, (E) serpin24, (F) serpin26, (G) toll3, (H) TC004646, (I) TC007763, (J) TC007858, (K) TC008806, (L) TC015479. See ‘Materials and methods’ for experimental details.
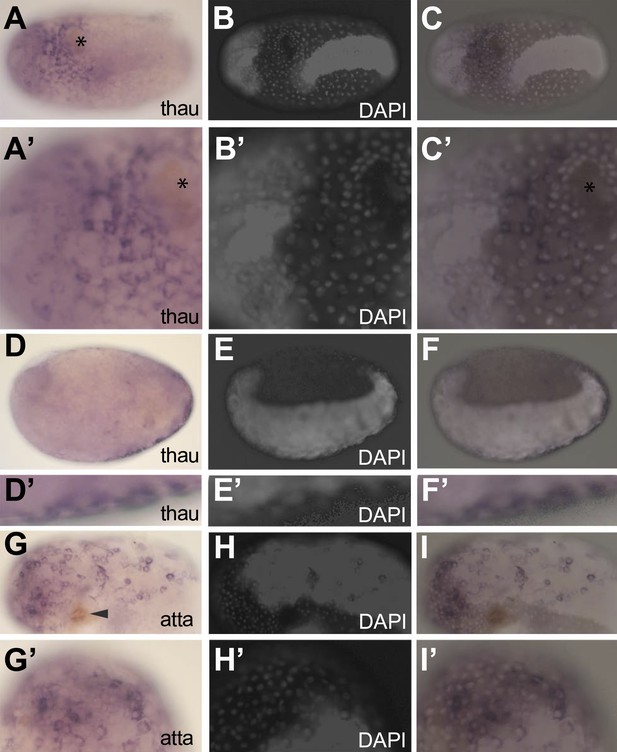
In situ hybridization showing expression of AMP genes in the serosa upon septic injury.
(A–F) Thaumatin1 in situ hybridization. (A) Superficial view. Thaumatin1 is expressed around the site of injury (asterix). Brown melanisation is observed around the site of injury. (A′) Magnification of the expression area shown in (A). Asterix marks the site of injury. (B) DAPI counterstaining of the same egg as in (A). The large polyploid serosal nuclei can be distinguished from the oversaturated DAPI signal from the germ-band. Head lobes to the left. (B′) magnification of (B). (C) Overlay of the in situ hybridization shown in A and the DAPI staining shown in (B). The thaumatin1 expression associates with the large polyploid serosal nuclei and is not found in the embryo proper. (C′) Magnification of the expression area shown in (C). (D) Focal plane through the egg. Thaumatin1 is expressed in a thin outer layer at the surface of the egg. (D′) Magnification of the expression area shown in (D). (E) DAPI staining of the same egg shown in (D). The embryo is brightly visible. Head to the left. (E′) Magnification of E. (F) Overlay of the in situ hybridization shown in D and the DAPI staining shown in (E). (F′) Magnification of the expression area. (G) Attacin1 in situ hybridization. Brown melanisation is visible around the site of injury (arrowhead). (G′) Magnification of the anterior region of the egg shown in (G). (H) DAPI staining of the same egg shown in G. The germ-band is brightly stained (head to the left) and the separate large serosal nuclei are visible. (H′) Magnification of the anterior of the egg shown in (H). (I) Overlay of the in situ hybridization shown in G and the DAPI staining shown in (H). Attacin1 is expressed in the large serosal cells covering the germ-band and is not expressed in the dense cells of the germ band. (I′) Magnification of the anterior of the egg shown in (I). The attacin1 staining associates with the large serosal nuclei.
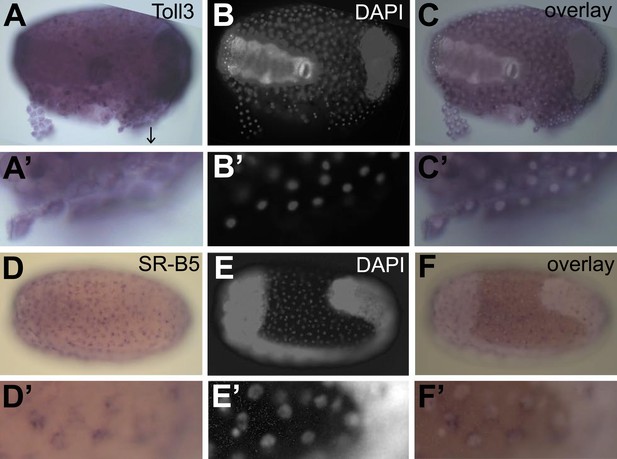
Constitutive expression of immune genes in the serosa.
(A–C) Toll3 in situ hybridization. (A) Toll3 is expressed in the flat and thin serosal cells (partly detached from the egg) but also in the germ rudiment (head lobes to the right). (A′) Magnification of the area indicated with an arrow in (A). (B) DAPI staining of the same egg shown in (A). The bright staining of the germ-band can be distinguished from the large nuclei of the serosa. (C) Overlay of the in situ hybridization shown in A and the DAPI staining shown in (B). (C′) Magnification of (C). Toll3 is expressed in cells of the serosa. (D–F) Scavenger receptor B5 in situ hybridization. (D) Scavenger receptor B5 shows expression in every serosal cell at the surface. (D′) Magnification of (D). (E) DAPI staining of the same egg shown in (D). The germ-band is brightly stained (head to the left) and the staining of the serosal nuclei is clearly visible when not overwhelmed by staining of the dense nuclei of the germ-band. (E′) Magnification of (E). The serosal nuclei are visible. Bright staining of the germ-band to the right. (F) Overlay of the in situ hybridization shown in D and the DAPI staining shown in (E). Scavenger receptor B5 expression follows the serosal nuclei and is not detected in the germ-band. (F′) Magnification of (F). Scavenger receptor B5 mRNA is detected around the large polyploid serosal nuclei and not around the dense nuclei of the germ rudiment.
Tables
Number of differentially expressed immune genes in Tribolium castaneum eggs
| Wild-type sterile injury | Wild-type septic injury | Control sterile injury | Control septic injury | Tc-zen1 sterile injury | Tc-zen1 septic injury | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Microbial recognition | ||||||||||||
| Extracellular signal transduction and modulation | ||||||||||||
| Intracellular transduction pathways (Toll/IMD/JNK/JAK-STAT) | ||||||||||||
| Execution/stress | ||||||||||||
| Total | ||||||||||||
-
Blue = induction, red = repression.
Antimicrobial properties of known and potential new antimicrobial peptides in Tribolium castaneum.
| Gene ID | Molecular weight (kDa) | Peptide length (AA) | Hydrophobic ratio | Net charge | Glycine content | Proline content | Fold change wild-type | Fold change control |
|---|---|---|---|---|---|---|---|---|
| Cecropin1/TC000499 | 3.67 | 31 | 58% | +5 | 6% | 0% | Inf | Inf |
| Cecropin3/TC000500 | 9.80 | 90 | 43% | +2 | 6% | 13% | Inf | 49x |
| attacin2/TC007738 | 15.80 | 145 | 37% | +7 | 12% | 4% | 3098x | 2190x |
| Coleoptericin1/TC005093 | 15.99 | 141 | 30% | −1 | 9% | 7% | 2392x | 18067x |
| Defensin2/TC010517 | 8.73 | 79 | 50% | +6 | 6% | 1% | 1183x | Inf |
| Defensin3/TC012469 | 9.42 | 83 | 50% | +7 | 3% | 1% | 908x | Inf |
| attacin1/TC007737 | 17.49 | 165 | 28% | +9 | 18% | 3% | 869x | 3696x |
| TC007858 | 20.14 | 182 | 35% | 0 | 11% | 3% | 484x | 54x |
| Defensin1/TC006250 | 14.91 | 132 | 46% | +11 | 4% | 3% | 187x | 1551x |
| TC011036 | 12.89 | 109 | 39% | +13 | 2% | 6% | 138x | 11x |
| Coleoptericin2/TC005096 | 15.96 | 141 | 30% | −1 | 9% | 7% | 91x | 227x |
| TC015479 | 13.00 | 120 | 42% | +6 | 5% | 1% | 80x | 26x |
| TC007763 | 16.87 | 158 | 37% | +4 | 6% | 17% | 47x | 67x |
| TC004646 | 15.04 | 135 | 34% | +2 | 7% | 7% | 40x | 29x |
| TC008806 | 15.83 | 142 | 33% | +2 | 10% | 2% | 31x | 37x |
| TC009336 | 13.50 | 137 | 30% | −4 | 39% | 2% | 15x | 7x |
| TC014565 | 20.73 | 176 | 38% | +17 | 2% | 2% | 14x | 9x |
| TC001030 | 14.62 | 137 | 29% | +9 | 10% | 12% | 9x | 16x |
| TC001784 | 13.54 | 150 | 27% | +7 | 43% | 2% | 8x | 6x |
| TC005478 | 13.70 | 122 | 45% | +10 | 4% | 1% | 6x | 7x |
| TC015612 | 20.32 | 182 | 36% | +7 | 6% | 6% | 6x | 2x |
| TC007901 | 7.25 | 64 | 25% | +5 | 7% | 10% | 6x | 5x |
| TC015304 | 19.30 | 180 | 38% | +2 | 6% | 9% | 5x | no hit |
| TC011733 | 11.89 | 106 | 46% | +3 | 2% | 0% | 5x | 5x |
| TC003374 | 12.22 | 124 | 61% | +3 | 1% | 9% | 2x | 9x |
| TC008557 | 17.82 | 172 | 31% | +2 | 18% | 0% | 3x | 5x |
| TC015754 | 15.69 | 140 | 34% | +5 | 4% | 7% | 2x | 2x |
| TC000435 | 11.84 | 105 | 37% | +5 | 5% | 0% | 2x | 2x |
| TC009096 | 12.84 | 111 | 16% | +16 | 9% | 6% | 2x | 2x |
-
In the table are known antimicrobial peptides and those proteins that show at least a twofold induction upon infection, they are smaller than 200 amino acids and are not negatively charged. TC009336 was included because of the high glycine content.
Differentially regulated immune genes in naive wild-type eggs compared to naive Tc-zen1 RNAi eggs
| Gene ID | Description | Fold change | FDR adjusted p-value | Gene ID | Description | Fold change | FDR adjusted p-value |
|---|---|---|---|---|---|---|---|
| Extracellular signal transduction and modulation | |||||||
| TC000247 | cSPH-H2 | 2.70 | <0.01 | TC005754 | serpin22 | 5.26 | <0.01 |
| TC000248 | cSPH-H3 | 4.11 | <0.01 | ||||
| TC000249 | cSPH-H4 | 5.16 | <0.01 | TC011718 | serpin27 | 1.62 | <0.01 |
| TC000740 | SPH-H17 | 9.28 | <0.01 | TC006726 | Spz4 | 3.00 | <0.01 |
| TC000829 | SPH-H18 | 8.26 | <0.01 | TC013304 | Spz5 | 122.56 | <0.01 |
| TC007026 | cSPH-H78 | 29.79 | <0.01 | Microbial recognition | |||
| TC012390 | SPH-H129 | 1.60 | <0.01 | TC002789 | PGRP-LA | 3.95 | 0.02 |
| TC000495 | cSP-P8 | 6.57 | <0.01 | TC014664 | TEP-B | 2.90 | 0.02 |
| TC000497 | cSP-P10 | 4.50 | <0.01 | TC005976 | PSH | 3.43 | <0.01 |
| TC000547 | SP-P13 | 2.41 | <0.01 | TC006978 | C-type lectin1 | 14.52 | <0.01 |
| TC000635 | SP-P16 | 2.54 | <0.01 | TC013911 | C-type lectin 13 | 18.21 | <0.01 |
| TC004160 | cSP-P44 | 9.79 | <0.01 | Toll-signalling pathway | |||
| TC004438 | Toll3 | 2.28 | <0.01 | ||||
| TC004635 | cSP-P53 | 51.56 | <0.01 | IMD-signalling pathway | |||
| TC005230 | cSP-P61 | 250.00 | <0.01 | TC014708 | NFAT | 2.01 | <0.01 |
| TC006033 | SP-P68 | 1.54 | <0.01 | Execution mechanisms | |||
| TC009090 | cSP-P91 | 2.80 | <0.01 | ||||
| TC009092 | cSP-P93 | 3.00 | <0.01 | TC005493 | Heme peroxidase 1 | 3.84 | <0.01 |
| TC009093 | cSP-P94 | 27.76 | <0.01 | TC015234 | Heme peroxidase 2 | 6.30 | <0.01 |
| TC013277 | cSP-P136 | 3.04 | <0.01 | ||||
| TC013415 | SP-P141 | 11.85 | <0.01 | TC015854 | Scavenger receptor-B2 | 1.91 | <0.01 |
| TC000760 | serpin1 | 5.29 | <0.01 | TC014946 | Scavenger receptor-B5 | 29.29 | <0.01 |
| TC005750 | serpin18 | 1.92 | <0.01 | TC000948 | Scavenger receptor-B6 | 163.90 | <0.01 |
| TC005752 | serpin20 | 2.30 | <0.01 | TC014954 | Scavenger receptor-B9 | 1.96 | <0.01 |
-
SP = serine protease; SPH = non-catalytic serine protease; cSP = clip-domain serine protease.
Additional files
-
Supplementary file 1
Summary statistics for Tribolium castaneum transcriptome sequencing analysis.
- https://doi.org/10.7554/eLife.04111.013
-
Supplementary file 2
Significantly differentially expressed immune genes in wild-type, control, and Tc-zen1 RNAi eggs.
- https://doi.org/10.7554/eLife.04111.014
-
Supplementary file 3
Primers used for RT-qPCR.
- https://doi.org/10.7554/eLife.04111.015
-
Supplementary file 4
DEseq output from wild-type eggs, sterile injury compared to naive eggs.
- https://doi.org/10.7554/eLife.04111.016
-
Supplementary file 5
DEseq output from wild-type eggs, septic injury compared to naive eggs.
- https://doi.org/10.7554/eLife.04111.017
-
Supplementary file 6
DEseq output from control RNAi eggs, sterile injury compared to naive eggs.
- https://doi.org/10.7554/eLife.04111.018
-
Supplementary file 7
DEseq output from control RNAi eggs, septic injury compared to naive eggs.
- https://doi.org/10.7554/eLife.04111.019
-
Supplementary file 8
DEseq output from Tc-zen1 RNAi eggs, sterile injury compared to naive eggs.
- https://doi.org/10.7554/eLife.04111.020
-
Supplementary file 9
DEseq output from Tc-zen1 RNAi eggs, septic injury compared to naive eggs.
- https://doi.org/10.7554/eLife.04111.021





