Crystal structure of PfRh5, an essential P. falciparum ligand for invasion of human erythrocytes
Figures
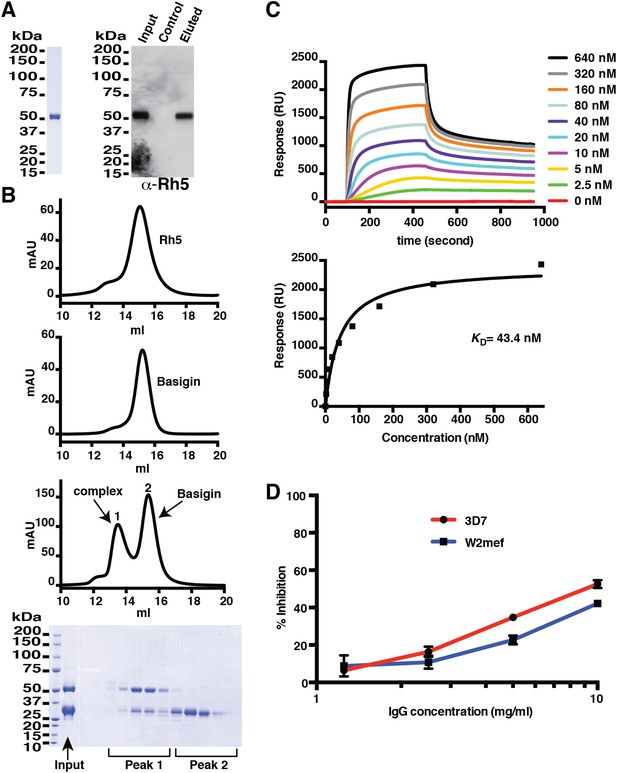
Production of functional recombinant PfRh5.
(A) Purified recombinant PfRh5 was analysed by SDS-PAGE analyses and by erythrocyte binding assays. (B) Formation of the PfRh5–basigin complex was monitored by size-exclusion chromatography. The chromatographic profiles are shown for PfRh5 (panel 1), basigin (panel 2), and the PfRh5-basigin complex (panel 3). The fractions eluted from the column in panel 3 were analysed by SDS-PAGE. (C) The binding affinity of the recombinant PfRh5 to human basigin was measured by SPR on Biacore 3000 with the basigin coupled to a sensor chip. (D) In vitro growth inhibition assays were performed to assess the abilities of the polyclonal antibodies to the recombinant PfRh5 in blocking P. falciparum parasite invasion into erythrocytes.
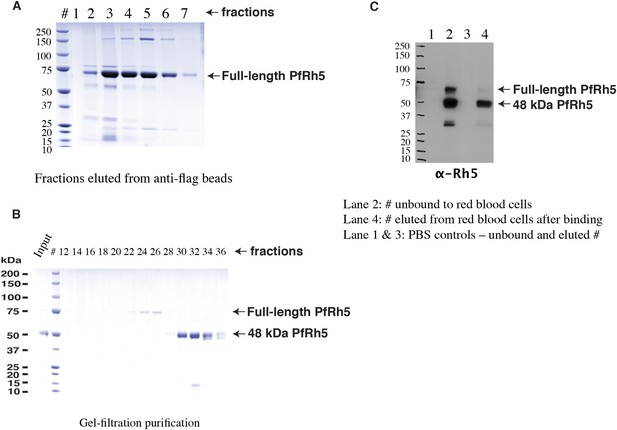
Production of full-length PfRh5.
(A) SDS-PAGE analyses of the FLAG-tagged recombinant full-length PfRh5 eluted from an anti-FLAG affinity column. (B) Size-exclusion chromatography analyses of the anti-FLAG bead affinity purified PfRh5. The fractions eluted from the affinity column were pooled, concentrated, and stored at 4°C for 2 days before the analyses. (C) Red blood cell binding assay with PfRh5 purified by anti-FLAG bead affinity chromatography. Both full-length protein and the breakdown fragments bound red blood cells with a 48-kDa species having the highest affinity.
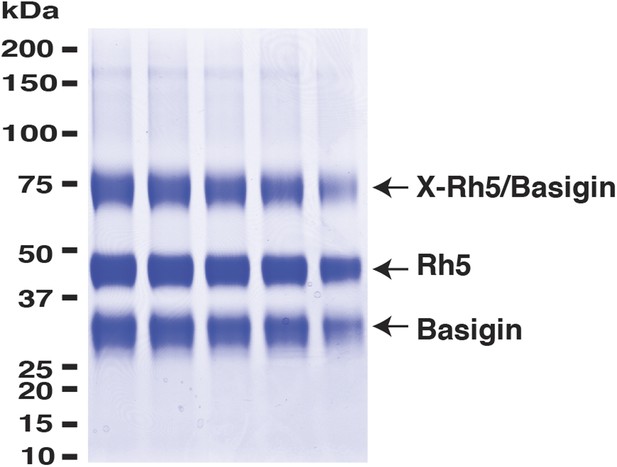
PfRh5 and human basigin form a 1:1 complex.
PfRh5-C and basigin were cross-linked with EDC in the presence of NHS. The mixture was then analysed on a SDS-PAGE gel and the band of the crosslinked complex was excised for trypsin/chymotrypsin digestion followed by mass spectrometric analysis.
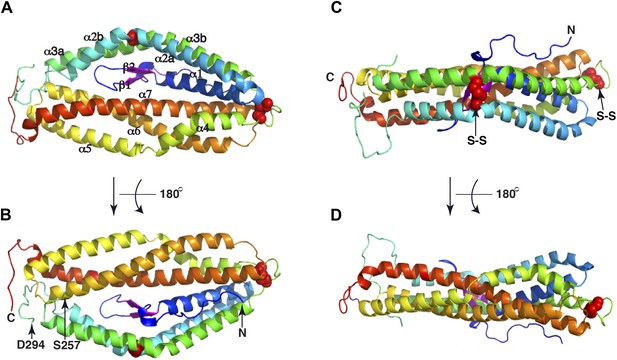
The crystal structure of PfRh5.
(A) Ribbon representation of the PfRh5 structure. Color scheme is rainbow (N-terminus: blue; C-terminus: red) except for the β-hairpin that is colored magenta for clarity. The cysteine residues that form disulfide bridges (Cys345–Cys351 and Cys224–Cys317) are shown as spheres. Helices α4, α5, α6, and α7 assemble as a triplet-helical coiled-coil domain running the length of the long axis of the molecule, helices α1, α2a, and α3b assemble to form a short triplet-helical bundle and helices α2b and α3a assemble to form a short two-helix coiled coil. (B) Ribbon representation of the PfRh5 structure viewed after a 180o rotation relative to that in (A). The residues between Ser257 and Asp294 are disordered. (C) Ribbon representation of the PfRh5 structure viewed from the side with the C-terminus on the left. The disulfide bridges are indicated with arrows. (D) Ribbon representation of the PfRh5 structure viewed after a 180o-rotation relative to that in (C).
-
Figure 2—source data 1
Data collection and refinement statistics of Rh5 and Rh5_KI.
- https://doi.org/10.7554/eLife.04187.007
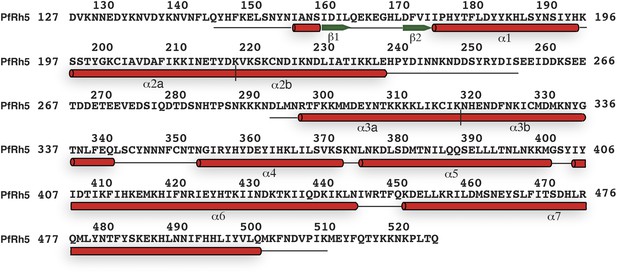
The secondary structure of PfRh5.
Density for the N-terminal residues Asp127–Leu145, loop residues Glu258–Asn293, and C-terminal residues Met512–Gln526 was not observed. The helices and β-strands are colored red and green respectively. The vertical line at residue 119 and 319 indicates the position of a helical break.
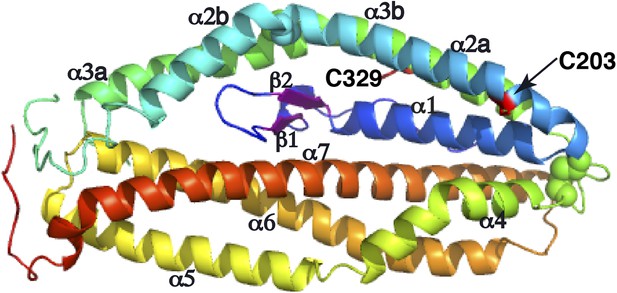
Two free cysteine residues (Cys203 on α2a and Cys329 on α3b) within the crystal structure of PfRh5.
The side chain of Cys329 is completely buried inside the three-dimensional fold of PfRh5 and the side chain of Cys203 is only partly exposed, consistent with the fact that no intermolecular disulfide bond was observed for the native and recombinant PfRh5.
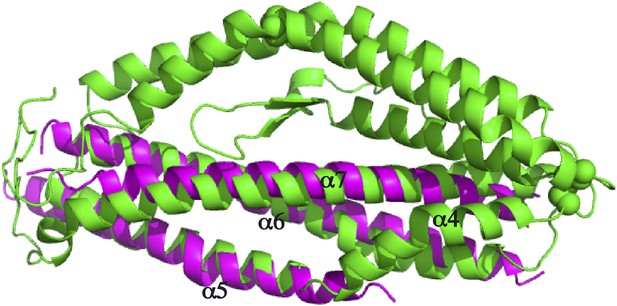
Superimposition of the PfRh5 structure with N-terminal coiled-coil domain of SipB.
The PfRh5 (green) coiled-coil helix bundle formed by helices α5, α6, and α7 has a very similar fold to the N-terminal coiled-coil domain of SipB (magenta).
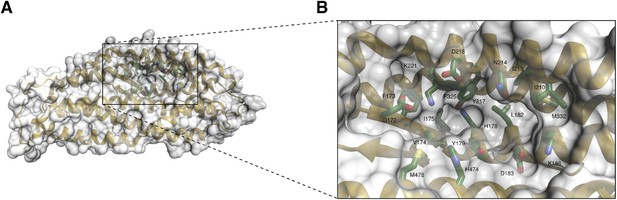
A unique pocket on the surface of the PfRh5 molecule.
(A) Location of the pocket on the surface formed by the β-hairpin, the triple-helical bundle, and the triple-helical coiled coil. (B) Close-up view of the pocket; key residues lining the pocket are labeled.
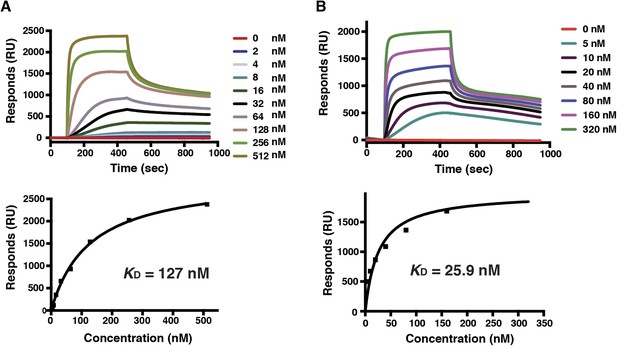
Binding of mutant PfRh5 to basigin.
(A) The binding affinity of the reduced and alkylated PfRh5 to basigin measured by SPR. (B) The binding affinity of the PfRh5 mutant, in which the disordered loop has been deleted, to basigin measured by SPR.





