Effective use of a horizontally-transferred pathway for dichloromethane catabolism requires post–transfer refinement
Figures
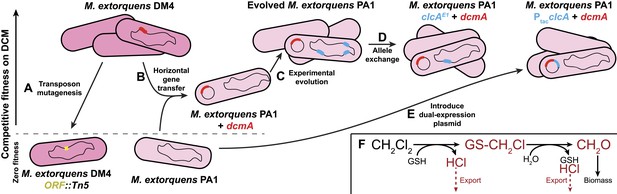
Experimental evolution recapitulates post–transfer optimization of a challenging catabolic pathway.
The natural isolate, DM4, grows well on dichloromethane. (A) Past efforts to explain the genetics of growth on DCM relied on gene knockouts (yellow) to identify important genes (Muller et al., 2011b). (B) We deliberately transferred the dcmA gene (red) into naïve recipient Methylobacterium strains, which then grew poorly on DCM (Michener et al., 2014). (C) Serial propagation on DCM selected for mutants with increased fitness on DCM. (D) Whole genome resequencing allowed us to identify these mutations (blue), and reconstructing the individual mutations in wild type cells verified that they were causal. We then worked backwards from the mutations to identify the stress that the mutations overcame. (E) Introducing a plasmid containing both the pathway (red) and a solution to the most common limiting stress (blue) allowed efficient growth on DCM without chromosomal modifications to the host. (F) The biochemistry of DCM dehalogenation produces several challenging compounds (red) that potentially limit growth on DCM.
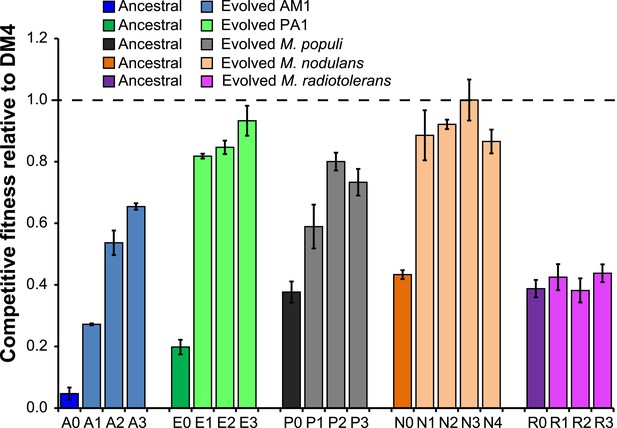
Evolved isolates from four of the five transconjugants have improved fitness relative to the ancestor.
A single clonal isolate was selected from each replicate population after 150 generations of growth on DCM. Each isolate was mixed with the reference strain, DM4ΔdcmA+pJM10, and grown on DCM to determine competitive fitness. Error bars show ±1 standard deviation, calculated from three biological replicates. The horizontal dashed line indicates equal fitness to the reference isolate, DM4.
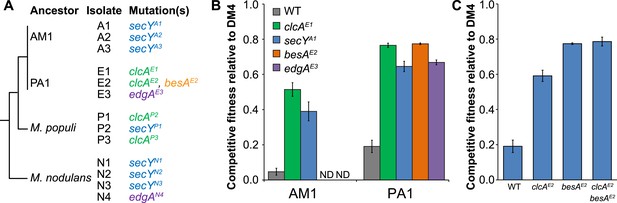
Each evolved isolate has a mutation in at least one of four loci.
(A) A combination of whole genome resequencing and targeted Sanger sequencing allowed the identification of mutations at four loci, encoding the protein translocase secY, the chloride/proton antiporter clcA, a hypothetical protein that we named edgA, and the bestrophin-homolog chloride channel besA. A simplified phylogenetic tree is shown at left (Michener et al., 2014). Mutation names indicate the genetic locus followed by the isolate name in which it has been detected. (B) Reconstruction of these single mutations in AM1 and PA1 demonstrated that each individual mutation was beneficial, even in a host in which the mutation was not observed (secY in PA1 and clcA in AM1). (C) In PA1, the double mutant besAE2/clcAE2 has similar fitness to the besAE2 single mutant when grown on DCM. During the evolution of population E2, the clcAE2 mutation fixed first, followed by the besAE2 mutation. Each of the reconstructed strains expresses dcmA from plasmid pJM10. Error bars show ±1 standard deviation, calculated from three biological replicates. N.D.: not determined.
-
Figure 3—source data 1
Mutations identified during experimental evolution. N.D.: not determined.
- https://doi.org/10.7554/eLife.04279.006
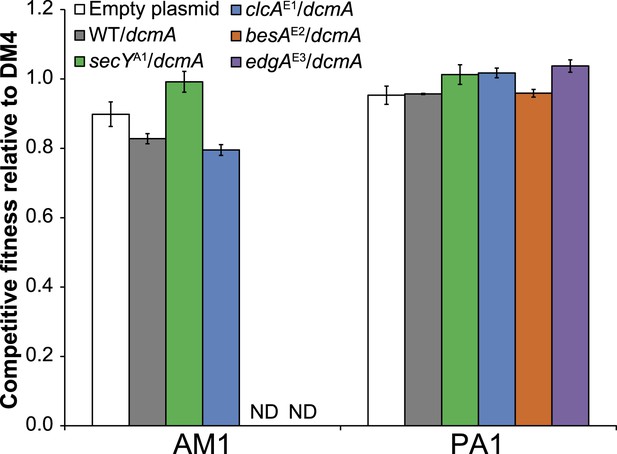
Mutations have relatively small fitness effects during growth on succinate compared to the benefits of threefold or more on DCM.
https://doi.org/10.7554/eLife.04279.007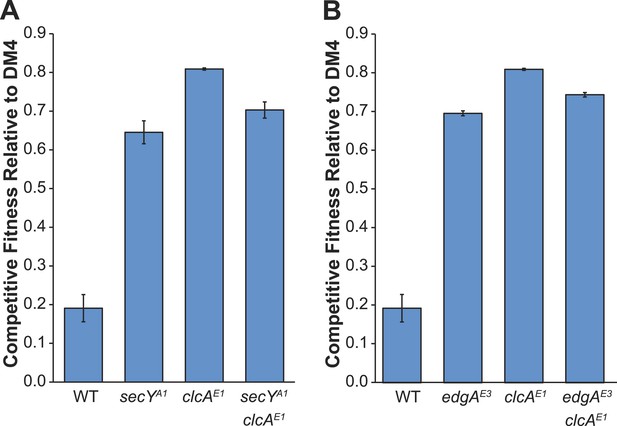
Comparison of competitive fitness of single and double mutants in M. extorquens PA1 explains evolutionary patterns.
All tested strains contain dcmA in pJM10. (A) The secYA1/clcAE1 PA1 double mutant is less fit than the clcAE1 single mutant during growth on DCM. (B) The edgAE3/clcAE1 PA1 double mutant is less fit than the clcAE1 single mutant during growth on DCM.
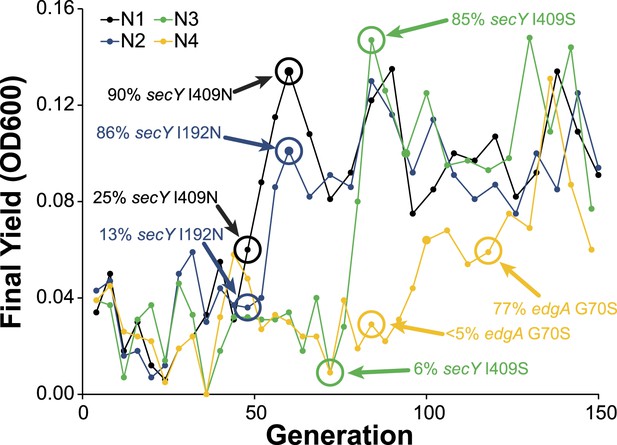
Allele frequency dynamics for M. nodulans populations.
For each of the four M. nodulans populations, the yield (as measured by OD600) was determined at the end of each serial culture. The near fixation of the indicated mutations, to secY and edgA, correspond to dramatic transitions in the culture yield.
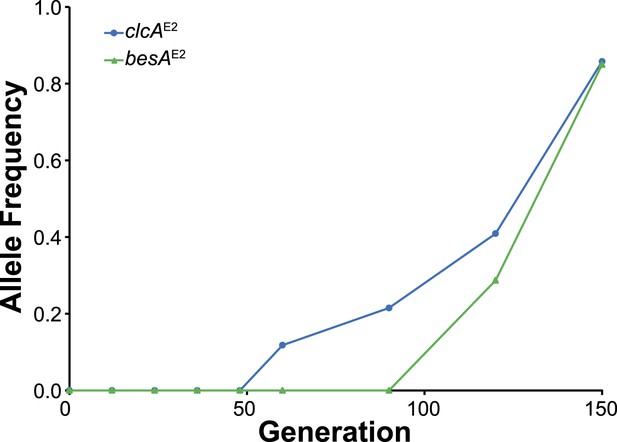
In population E2, the clcAE2 mutation fixed first, followed by besAE2.
As shown in Figure 3C, the besAE2 mutation was moderately beneficial in the clcAE2 background, while the reverse was not true.
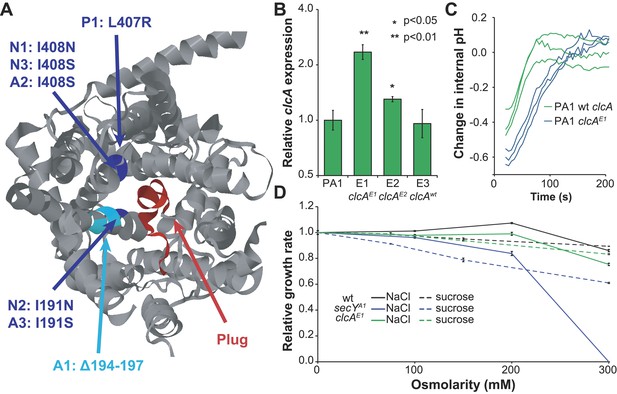
Mutations in secY and clcA are predicted to increase chloride export.
(A) When mapped to the crystal structure of SecY from Methanocaldococcus jannaschii (Van den Berg et al., 2004), the seven mutations to secY cluster in a small region of the protein structure (blue and cyan) at the interface between the channel and plug (red) that is associated with leakage of small anions. (B) Mutations to the clcA promoter in isolates E1 and E2 lead to significantly increased transcription of clcA, as measured by qRT-PCR (one tailed Welch's t-test with two degrees of freedom). Error bars show ±1 standard deviation, calculated from three biological replicates. (C) Adding DCM to strains that overexpress clcA leads to a larger decrease in internal pH. Strains contain pJM40, expressing both dcmA and a fluorescent pH biosensor. The strain with a mutated clcA promoter (blue) overexpresses ClcA relative to the wild type strain (green). At t = 0, DCM was added to a final concentration of 10 mM. After an approximate delay of 20 s, the culture was continuously sampled with a flow cytometer to determine the dynamics of the internal pH. Both strains showed a transient decrease in pH, but the decrease is larger for the clcA overexpression strain (p = 0.036, one-tailed Welch's t-test with two degrees of freedom). (D) Mutations to secY, but not to clcA, increase chloride sensitivity. Growth rates of three PA1 strains (wild type, secYA1, and clcAE1) were measured using an automated assay system. The osmolarity of the medium was varied through the addition of sucrose or sodium chloride. The secYA1 mutant did not grow at the highest chloride concentration.
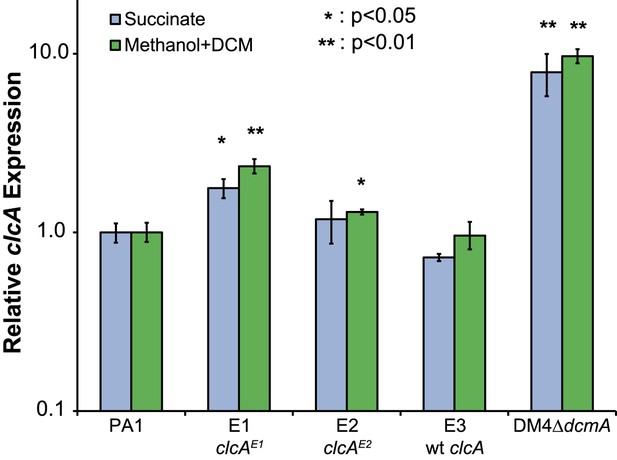
Relative clcA expression is independent of the growth substrate.
All five strains were grown to mid-log phase in M-PIPES containing either 5 mM DCM plus 0.075 mM methanol (green) or 3.5 mM succinate (blue). For each growth substrate, clcA expression was measured by qRT-PCR and compared to wild type PA1 under the same conditions. Relative clcA expression across different strains was similar for both growth conditions. Error bars show ±1 standard deviation, calculated from three biological replicates. p values are calculated using a one-tailed Welch's t-test with two degrees of freedom.
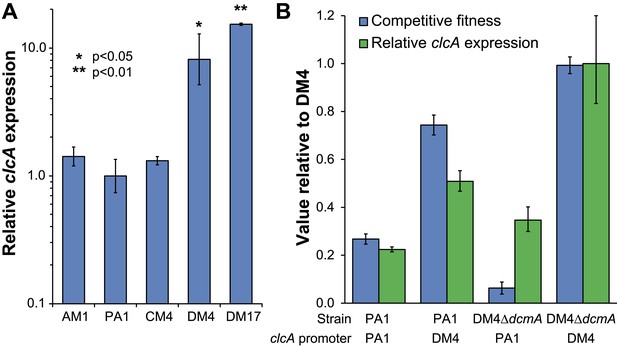
DM4 requires clcA overexpression to grow on DCM.
(A) The environmental DCM-degrading isolates, DM4 and DM17, overexpress clcA relative to three other strains of M. extorquens (one tailed Welch's t-test with two degrees of freedom). Error bars show ±1 standard deviation, calculated from three biological replicates. (B) Growth on DCM in DM4 and PA1 depends on the clcA promoter but not the strain background. A 140-bp region upstream of clcA (the ‘clcA promoter’) was swapped between PA1 and DM4. Regardless of the genetic background, strains with the DM4 clcA promoter overexpressed clcA and were more fit on DCM compared to strains with the PA1 clcA promoter. All strains expressed dcmA from plasmid pJM10. Error bars show ±1 standard deviation, calculated from three biological replicates.
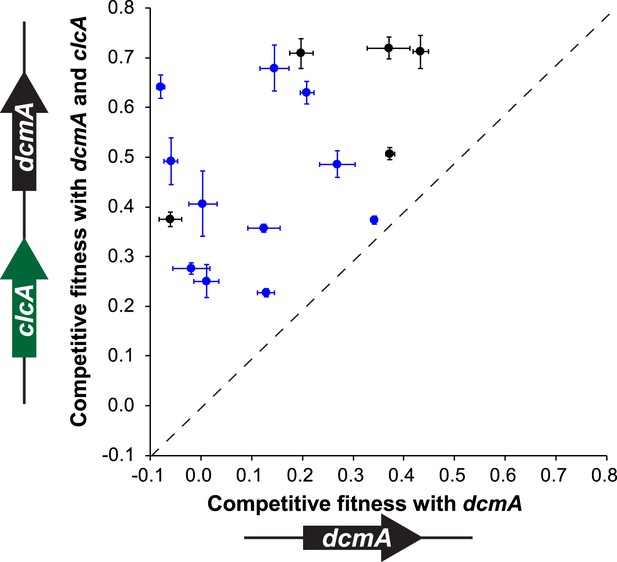
A mobile genetic element containing both dcmA and clcA allows a diverse array of Methylobacterium isolates to grow efficiently on DCM.
Plasmids containing either dcmA (x-axis) or dcmA and clcA (y-axis) were introduced into 16 different Methylobacterium strains, including the five strains used to found the evolved populations (black) and 11 that were new (blue). Each transconjugant was tested against the reference, DM4ΔdcmA+pJM10, to determine its competitive fitness. Note that negative fitness values indicate net death over the course of the competition. The dashed line indicates equal fitness of the two transconjugants, as would be expected if clcA expression were neutral. Error bars show ±1 standard deviation, calculated from three biological replicates.
-
Figure 6—source data 1
Environmental Methylobacterium strains characterized with the dual expression plasmid.
Closest relative was determined based on the nearest BLAST hit to the 16S rRNA sequence of the isolate.
- https://doi.org/10.7554/eLife.04279.016
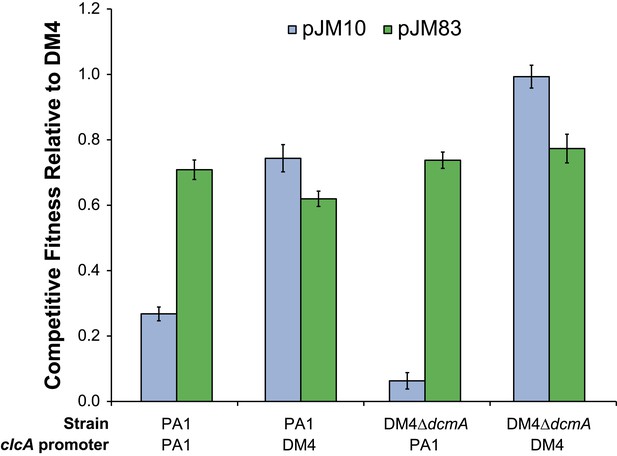
Overexpression of clcA, either by the DM4 clcA promoter or the pJM83 clcA/dcmA dual expression plasmid, confers high fitness on DCM.
Replacing the clcA promoter in DM4 ΔdcmA with the clcA promoter from PA1 severely reduces fitness on DCM. However, this fitness defect can largely be compensated through clcA overexpression by the introduction of the pJM83 dcmA/clcA dual expression plasmid. Error bars show ±1 standard deviation, calculated from three biological replicates.
Additional files
-
Supplementary file 1
Strains and plasmids used in this study.
- https://doi.org/10.7554/eLife.04279.017





