Plant defense phenotypes determine the consequences of volatile emission for individuals and neighbors
Figures
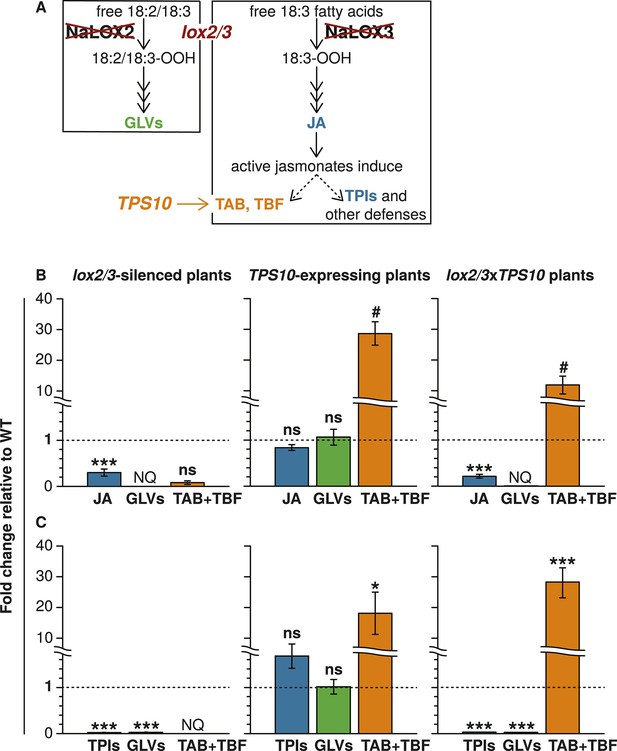
The lox2/3 and TPS10 transgenic constructs, alone and in combination (lox2/3xTPS10), independently alter herbivore-induced plant defenses and sesquiterpene HIPVs.
(A) The endogenous lipoxygenase genes LOX2 and LOX3 and the Z. mays sesquiterpene synthase gene TPS10 (TPS10) were manipulated in transgenic lines of N. attenuata in order to uncouple the production of two HIPVs—the sesquiterpenes (E)-α-bergamotene (TAB) and (E)-β-farnesene (TBF)—from jasmonate-mediated direct defenses and other volatiles. LOX2 and LOX3 provide fatty acid hydroperoxides from 18:2 and 18:3 fatty acids (18:2/18:3-OOH) for the synthesis of green leaf volatiles (GLVs) or jasmonic acid (JA) and the jasmonate-derived hormones, respectively. GLVs are released in response to herbivore damage, and jasmonates regulate the production of most anti-herbivore defenses, including trypsin protease inhibitors (TPIs), and other HIPVs. LOX2 and LOX3 were transgenically silenced using an inverted repeat (ir) construct specifically silencing both (lox2/3). Jasmonate-regulated volatiles in N. attenuata are mostly sesquiterpenes, of which the most prevalent is TAB, and include trace amounts of the biosynthetically related sesquiterpene TBF. The Z. mays sesquiterpene synthase TPS10 (Köllner et al., 2009), which produces TAB and TBF as its main products, was ectopically over-expressed in N. attenuata plants under control of a 35S promoter (TPS10) to uncouple the emission of these volatiles from endogenous jasmonate signaling. (B and C) Plants with lox2/3 and TPS10 constructs, and a cross with both constructs having the transgenes in a hemizygous state (lox2/3xTPS10) had the expected phenotypes both in glasshouse and field experiments. For each compound or group of compounds, the mean WT value was set to 1 (indicated by dashed lines), and all values were divided by the WT mean resulting in mean fold changes ±SEM. A complete description of TPS10 plants including the demonstration of a single transgene insertion and WT levels of non-target metabolites is provided in Schuman et al. (2014), and additional information for lox2/3, TPS10, and lox2/3xTPS10 plants (single transgene insertion for lox2/3, accumulation of target gene transcripts) is given in Appendix 1. (B) Glasshouse-grown plants were treated with wounding and M. sexta OS, and JA, GLVs, TAB and TBF were analyzed at the time of peak accumulation; n = 4. For lox2/3 and lox2/3xTPS10, GLVs were not quantifiable (NQ) by GC–MS due to inconsistently detected or no detected signals. ***p<0.001 in Tukey HSD tests following a significant one-way ANOVA (p<0.001) of WT, lox2/3, and lox2/3xTPS10; # plants with the TPS10 construct emit significantly more TAB + TBF (Holm-Bonferroni-corrected p<0.01 in a Wilcoxon rank sum test of WT and lox2/3 vs lox2/3xTPS10 and TPS10); ns, not significant. TBF was not detectable in lox2/3 or WT samples. For absolute amounts of JA and other phytohormones, GLVs, TAB and TBF measured in glasshouse samples, see Figure 1—source data 1, 2. (C) TPI activity, total detectable GLVs, and total TAB and TBF measured in frozen leaf samples harvested from field-grown plants at the end of experimental season two (June 28th), after plants had accumulated damage from naturally occurring herbivores. TPIs (n = 11–24) were slightly elevated in field samples of TPS10 plants, but TPI activity did not differ from WT plants in glasshouse samples from two independent TPS10 lines including the line used for this study (Schuman et al., 2014). Total GLVs, TAB, and TBF were quantified by GC–MS in hexane extracts from the same tissue samples used to measure TPI activity, n = 9–22 (a few samples had insufficient tissue for the analysis). Neither TAB nor TBF could be detected in lox2/3 samples (NQ); TBF was also not detectable in WT samples. *Corrected p<0.05, ***corrected p<0.001 in pairwise Wilcoxon rank sum tests following significant (corrected p<0.05) Kruskal–Wallis tests across all genotypes for each category; p-values were corrected for multiple testing using the Holm-Bonferroni method; ns, not significant. For absolute amounts of TPIs, GLVs, TAB and TBF, and non-target volatiles measured in field-collected tissue samples, see Figure 1—source data 3; emission of GLVs, TAB, TBF, and non-target volatiles from field-grown plants is given in Appendix 2.
-
Figure 1—source data 1
Absolute values for JA and GLVs shown as fold-changes in Figure 1B in lox2/3 and lox2/3xTPS10 plants, as well as JA-Ile, individual GLVs, and leaf areas for headspace collection (mean ± SEM); n = 4.
Data are from a single glasshouse experiment. Values for TPS10 (line 10–3) are reported in Schuman et al. (2014).
- https://doi.org/10.7554/eLife.04490.004
-
Figure 1—source data 2
Absolute values for TAB and TBF shown as fold-changes in Figure 1B, and leaf areas for headspace collection (mean ± SEM); n = 4.
Data are from a single glasshouse experiment.
- https://doi.org/10.7554/eLife.04490.005
-
Figure 1—source data 3
Absolute values for analytes shown as fold-changes in Figure 1C (mean ± SEM).
Data are from tissue samples of several pooled leaves per plant from field-grown plants, taken at the end of experimental season two (June 28th).
- https://doi.org/10.7554/eLife.04490.006
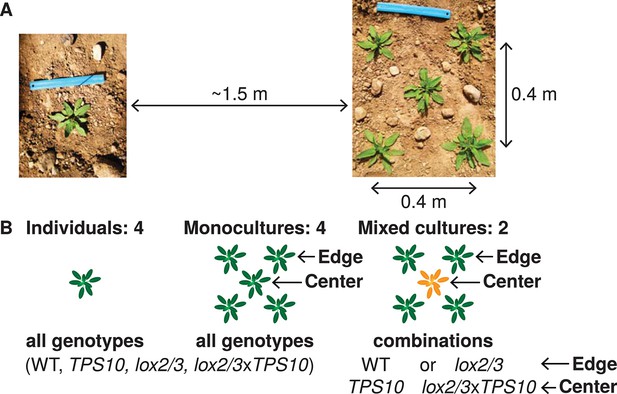
Field experiments designed to measure genotype-by-population effects of TPS10 volatiles in well-defended (WT, TPS10) or poorly defended (lox2/3, lox2/3xTPS10) plants.
Two similar experiments were conducted in two consecutive field seasons. (A) Each genotype was planted as individuals and in five-plant populations (see Figure 2—figure supplement 1). Each individual or population was planted ca. 1.5 m from the nearest neighboring individual/population, measured perimeter-to-perimeter. Populations consisted of five plants arranged as shown in a 0.4 × 0.4 m square. (B) Individual and population types: monocultures and, additionally, mixed cultures were planted in which a TPS10 plant (TPS10 or lox2/3xTPS10) was surrounded by four plants of the same level of jasmonate-mediated defense and GLVs (WT or lox2/3). Replicates (n = 12 in season one, 15 in season two of each individual or population type) were arranged in a blocked design (see Figure 2—figure supplement 2): blocks consisting of one replicate of each type (all six randomly-ordered populations followed by all four randomly-ordered individuals) were staggered such that consecutive blocks were not aligned, and thus populations and individuals were also interspersed. Random order was modified only when necessary to ensure that no two replicate individuals/populations were placed next to each other vertically, horizontally, or diagonally. This planting design reflects typical distributions in native populations of Nicotiana attenuata (see Figure 2—figure supplement 3), particularly before 1995, after which an invasive brome grass fueled a dramatic increase in fire sizes and consequently N. attenuata population sizes.

Photographs of the field experiment in season one.
(A) Photograph of a closed-headspace volatile trapping showing ca. 25% of the experiment on May 7th of season one. (B) Photograph showing a similar portion of the experiment on May 23rd of season one.
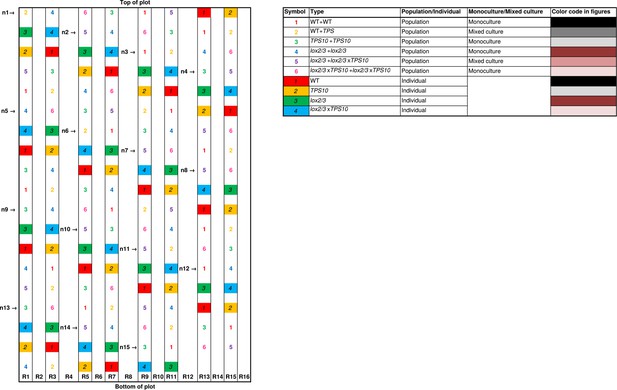
Layout in experimental season two, and color codes.
R, row of earth between two irrigation lines; not to scale.

Example of plant distribution in a native N. attenuata population, photographed in 2004.
Reprinted with permission from Danny Kessler, Copyright 2004. All rights reserved.
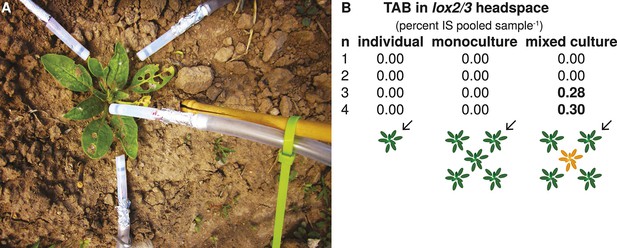
The TPS10 product TAB can be detected in the open headspace around lox2/3 plants in mixed cultures containing a lox2/3xTPS10 plant.
(A) An ‘open headspace’ trapping was conducted during season two (May 16th) with rosette-stage individuals and edge plants as shown, using activated charcoal filters shielded from ozone and UV by MnO2-coated copper ozone scrubbers (black strips in Teflon tubes pointed at plant) and aluminum foil, respectively. The headspace was sampled during the day, when sesquiterpenes are most abundant (see Appendix 2). Eluents from all four filters were combined and analyzed using highly sensitive GCxGC-ToF analysis; n = 4 plants. (B) The TPS10 product trans-α-bergamotene (TAB) could be detected in the open headspace of two of four lox2/3 plants at the edges of mixed cultures containing a lox2/3xTPS10 plant, but not in lox2/3 individuals or lox2/3 plants in monocultures. Peak areas were normalized to the internal standard (IS) peak. Arrows indicate the plant that was sampled.
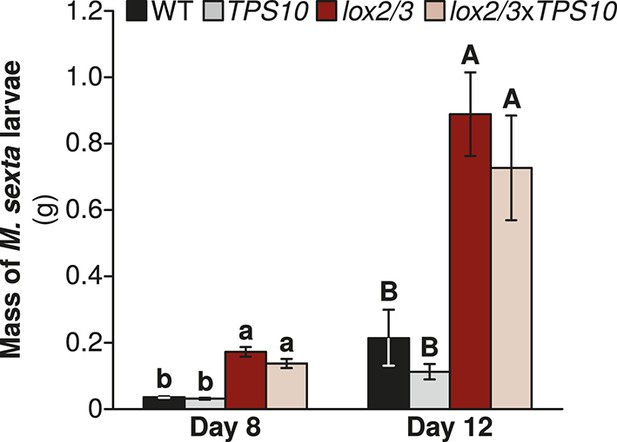
Manduca sexta larvae grow larger on LOX2/3-deficient plants, regardless of TPS10 expression, in a glasshouse experiment.
Data are shown as mean ± SEM; n = 15–19 larvae on day 8 and 11–16 larvae on day 12. One M. sexta neonate per plant (starting n = 25 larvae) was placed immediately after hatching on the youngest rosette leaf of an elongated plant. Larvae were weighed at the third and fourth instars (of 5 total), corresponding to days 8 and 12, after which larvae become mobile between plants. a,b/A,B Different letters indicate significant differences (corrected p<0.001) in larval mass on different plant genotypes within each day, in Wilcoxon rank sum tests following significant (corrected p<0.001) Kruskal–Wallis tests for each day (WT vs lox2/3 day 8, W17,19 = 310, corrected p<0.001, day 12, W15,15 = 217, corrected p<0.001; TPS10 vs lox2/3xTPS10, day 8, W15,20 = 290, corrected p<0.001, day 12, W11,14 = 143, corrected p<0.001; WT vs TPS10 day 8, W17,15 = 95, corrected p=0.227, day 12, W15,11 = 56, corrected p=0.361; lox2/3 vs lox2/3xTPS10 day 8, W19,20 = 249, corrected p=0.201, day 12, W16,14 = 134, corrected p=0.377). p-values were corrected for multiple testing using the Holm-Bonferroni method.
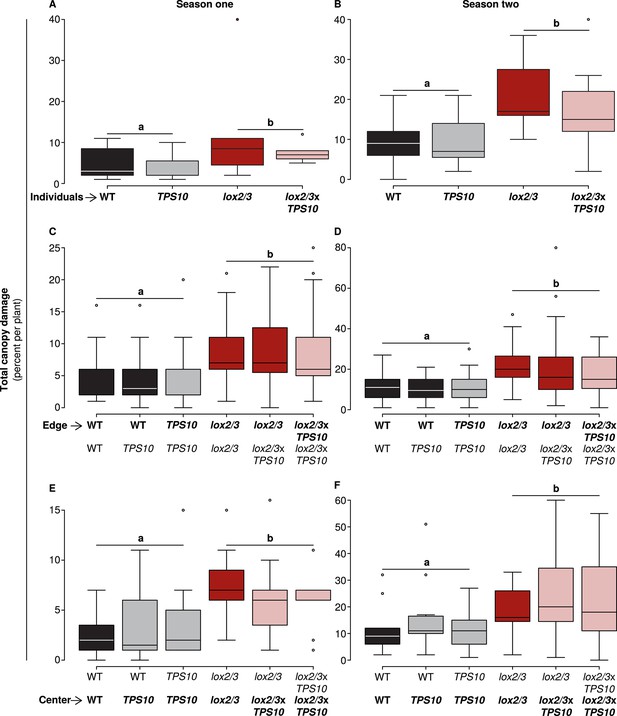
Only LOX2/3 and not TPS10, plant position, or population type determined total foliar damage from herbivores over two consecutive field seasons.
Note differences in y-axis scale. Data were collected on June 9th in season one (left panels), and on June 2nd in season two (right panels) from plants at all different positions in the experiment: individual plants (A–B), and plants at the edges (C–D) or at the centers (E–F) of populations. Black bars denote WT, grey bars TPS10, red bars lox2/3, and pink bars lox2/3xTPS10 plants. Total canopy damage reflected the trends in canopy damage caused by individual herbivores (Figure 5—source data 1, 2). Total canopy damage in season one was low (left panels), ranging from 5–10% on lox2/3 and lox2/3xTPS10 plants, and only 2–5% on WT and TPS10 plants. Damage levels were about twice as high in season two (right panels), but showed the same relative pattern, with LOX2/3-deficient plans having more damage. Damage levels within a season were similar for plants in different positions (p>0.07, see Appendix 3). a,b Different letters indicate significant differences (p<0.05) between LOX2/3-expressing and LOX2/3-deficient plants (WT, TPS10 v. lox2/3, lox2/3xTPS10) in minimal ANOVA or linear mixed-effects models on arcsin-transformed data. For individuals and center plants n = 7–13, and for edge plants n = 16–48 (up to 4 per population; the blocking effect was accounted for by a random factor in statistical analysis); exact replicate numbers are given in Figure 5—source data 1, 2. There were no significant differences in damage between plants differing only in TPS10 expression (p>0.4), plants in mono- vs mixed cultures (p>0.5), or plants of the same genotype at different positions (individual, edge, or center, p>0.07). Statistical models are given in Appendix 3.
-
Figure 5—source data 1
Total canopy damage from individual herbivores or groups of herbivores in experimental season one, corresponding to data shown in Figure 5A (mean ± SEM).
- https://doi.org/10.7554/eLife.04490.014
-
Figure 5—source data 2
Total canopy damage from individual herbivores or groups of herbivores in experimental season two, corresponding to data shown in Figure 5B (mean ± SEM).
- https://doi.org/10.7554/eLife.04490.015
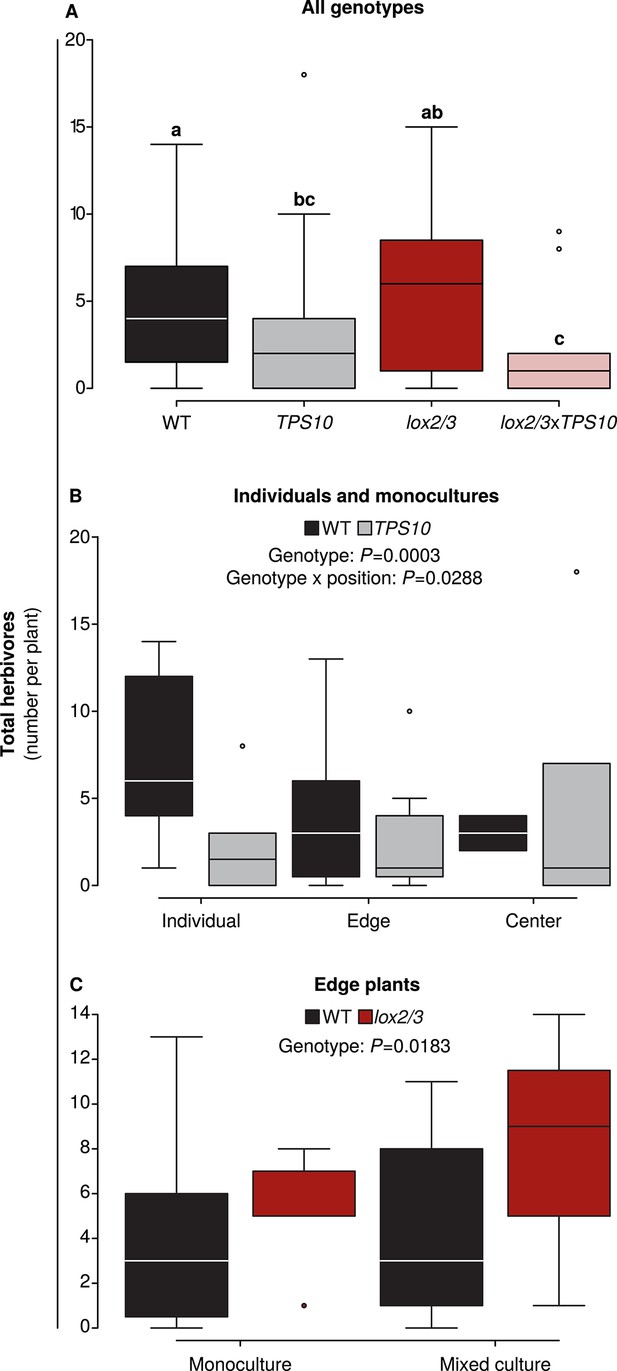
Foliar herbivore abundance is determined by TPS10 expression, lox2/3 expression, and plant position.
Data were collected on June 26th in season two, when herbivores and predators were more abundant (see Figure 5). Black denotes WT, grey TPS10, red lox2/3, and pink lox2/3xTPS10 plants. Counts for individual herbivores and predators are given in Figure 6—source data 1, 2. (A) Herbivore counts on focal plants of each genotype revealed fewer herbivores present on plants expressing TPS10 (n given in Figure 6—source data 1). a,b Different letters indicate significant differences (corrected p<0.05) between genotypes in Tukey contrasts following significant differences in a generalized linear mixed-effects model (see Appendix 3). (B) For WT and TPS10 individuals or plants in monocultures (n given in Figure 6—source data 2), there was a significant effect (p<0.05) of plant genotype (z = −3.662, p=0.0003) and an interaction of genotype with position (TPS10 by center vs individual position, z = 2.186, p=0.0288, generalized linear mixed-effects model in Appendix 3). (C) The presence of a TPS10-expressing neighbor in mixed populations did not significantly affect herbivore abundance on WT or lox2/3 edge plants, but there was a significant difference between WT and lox2/3 genotypes at the edges of populations (n given in Figure 6—source data 2, z = 2.358, p=0.0183, generalized linear model in Appendix 3).
-
Figure 6—source data 2
Total and median numbers of herbivores counted on focal plants of different genotypes on June 26th in season two, broken down into different population types and plant locations for WT and TPS10 plants.
Replicate numbers of lox2/3 and lox2/3xTPS10 plants were not sufficient for this analysis (n ≤ 3 per population type and position).
- https://doi.org/10.7554/eLife.04490.017
-
Figure 6—source data 1
Total and median numbers of herbivores and predators counted on focal plants of different genotypes on June 26th in season two.
- https://doi.org/10.7554/eLife.04490.018
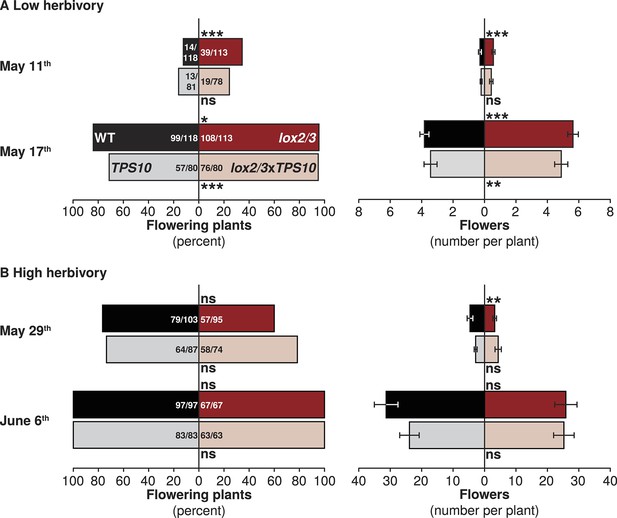
LOX2/3 deficiency accelerated flowering under low herbivory.
Flower production was monitored twice per season: once shortly after plants began to flower and again 1 week later, when most or all plants were flowering. Bars indicate either total percentage of plants of each genotype which were flowering (left panels), or the mean number of flowers per plant (right panels). WT and TPS10 (black and grey bars) are shown opposite lox2/3 and lox2/3xTPS10 (red and pink bars). Ratios in bars indicate numbers of flowering/total plants monitored; a few plants were not included in counts which had recently lost their main stem to herbivores and had not yet re-grown. (A) In season one, when plants suffered less damage from herbivores (Figure 5), plants with the lox2/3 silencing construct produced more flowers earlier: asterisks indicate pairwise differences between genotypes differing only in the lox2/3 silencing construct. ***Corrected p<0.001, **corrected p<0.01, *corrected p<0.05, or no significant difference (ns) in G-tests (percentage flowering) or in Wilcoxon rank sum tests (flower number, WT vs lox2/3: May 11th, W120,120 = 8582, p=0.0002; May 17th, W120,119 = 5043, p<0.0001; TPS10 vs lox2/3xTPS10: May 11th, W84,84 = 3801, p=0.2063; May 17th, W84,84 = 2520, p=0.0027). (B) In season two, during which plants received more damage from herbivores (Figure 5), flowering was monitored at later dates: the first timepoint in (B) is comparable to the second timepoint in (A) in terms of the proportion of plants flowering and the average number of flowers per plant (note difference in scale). In season two, LOX2/3 deficiency rather decreased early flower numbers in the comparison of WT vs lox2/3 on May 29th, and did not increase flower numbers at either measurement (WT vs lox2/3: May 29th, W103,95 = 3860.5, p=0.0091; June 6th, W97,69 = 2940.5, p=0.3018; TPS10 vs lox2/3xTPS10: May 29th, W87,74 = 3646.5, p=0.1417; June 6th, W83,63 = 2695, p=0.7518). For a complete analysis of the effects of LOX2/3 deficiency and TPS10 expression, plant position, and population type on flowering and plant size, see source data file for Figure 8.
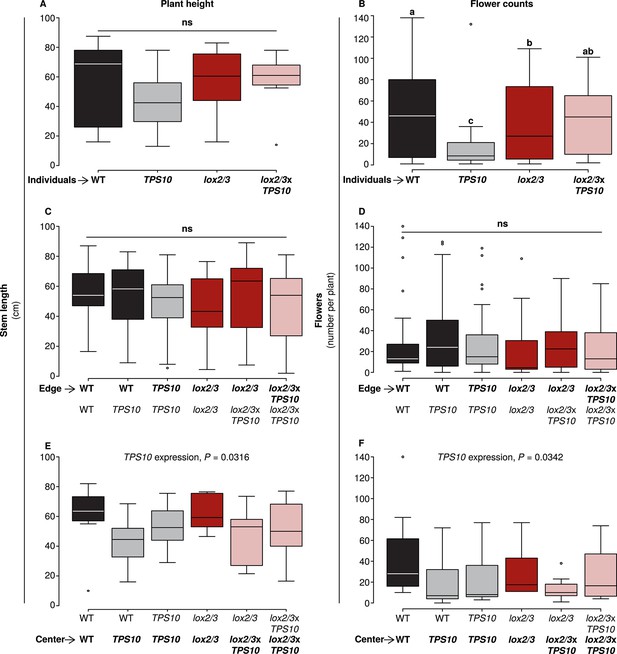
TPS10 expression reduced flower production under high herbivory.
Plant size as measured by stem length (left panels), and reproduction as measured by flower production (right panels); n given in Figure 8—source data 1. Data were collected on June 6th in season two from plants at all different positions in the experiment: individual plants (A–B), and plants at the edges (C–D) or at the centers (E–F) of populations. Black bars denote WT, grey bars TPS10, red bars lox2/3, and pink bars lox2/3xTPS10 plants. Plant height differed little, with only a small negative effect of TPS10 expression for center plants (E), which was more pronounced in mixed- than monocultures, indicating a slight competitive disadvantage for these plants. Individual plants differed much more than plants in populations in terms of flower production, with TPS10-expressing plants at a disadvantage (B); this effect was greatly reduced in populations.a,b Different letters indicate significant differences (corrected p<0.05) in Tukey post-hoc tests on plant genotype following significant effects in a generalized linear model, or generalized linear mixed-effects model (C–D) with genotype as a factor; these p-values were corrected using the Holm-Bonferroni correction for multiple testing as the same data were also tested for effects of LOX2/3 deficiency and TPS10 expression (see Figure 8—source data 1). There was no difference in flower production for edge plants, but a small negative effect of TPS10 expression on flower production in center plants (F) corresponding to the slight reduction in height of these plants (E). Statistical models are given in Figure 8—source data 1.
-
Figure 8—source data 1
The same statistical approach described in Appendix 3 for Figure 5 was used for plant size and reproduction data from May 17th in season 1, and June 6th in season two (the most complete measurements, closest to the date of herbivore damage screens, from each season).
It should be kept in mind that plants in the season one analysis were at an earlier stage of growth and flowering, for example, had about 10% as many flowers as plants in the season 2 analysis; for an analysis of flowering at similar stages, see Figure 7. Significant p-values and factors are given in bold, while p-values and factors marked in grey and in bold are marginal, but are required in the minimal model because their removal resulted in poorer (higher) AIC values. Other non-significant factors remained in models because they were the least insignificant factor (if only one factor remained), or because they are dependent on other factors which are significant or required.
- https://doi.org/10.7554/eLife.04490.021
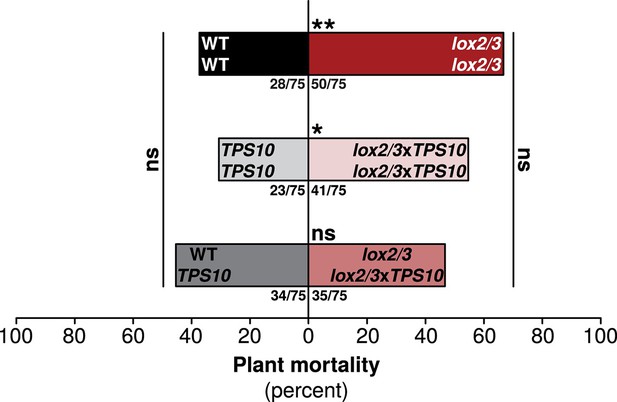
One TPS10-expressing plant per population counteracts a mortality increase in LOX2/3-deficient populations.
Bars indicate total percentage mortality for all plants in each population type, and ratios in bars indicate numbers of dead/total plants (n = 75). WT and TPS10 populations (left, black, and grey bars) are compared to the equivalent lox2/3 and lox2/3xTPS10 populations (right, red, and pink bars) differing only in the lox2/3 silencing construct. LOX2/3-deficient plants had higher mortality in monocultures, regardless of whether they also expressed TPS10 (**WT mono v. lox2/3 mono, corrected p<0.01; *TPS10 mono v. lox2/3xTPS10 mono, corrected p<0.05 in G-tests; p-values were corrected for multiple testing using the Holm-Bonferroni method). However, the mortality of plants in mixed lox2/3 + lox2/3xTPS10 populations is similar to that of the plants in mixed WT + TPS10 populations (ns, not significant). There was no significant difference in mortality among mono- and mixed cultures of WT and TPS10 plants (corrected p>0.3), or of lox2/3 and lox2/3xTPS10 plants, although there was a marginal difference between lox2/3 monocultures and lox2/3 + lox2/3xTPS10 mixed cultures (corrected p=0.091). In addition, a health index was assigned to each plant (discussed in text); this index was correlated with several plant growth and reproduction parameters (see Figure 9—figure supplement 1). Data were collected on June 29th at the end of experimental season two; in season one, all plants experienced much lower herbivory (Figure 5) and negligible mortality (0–5%).
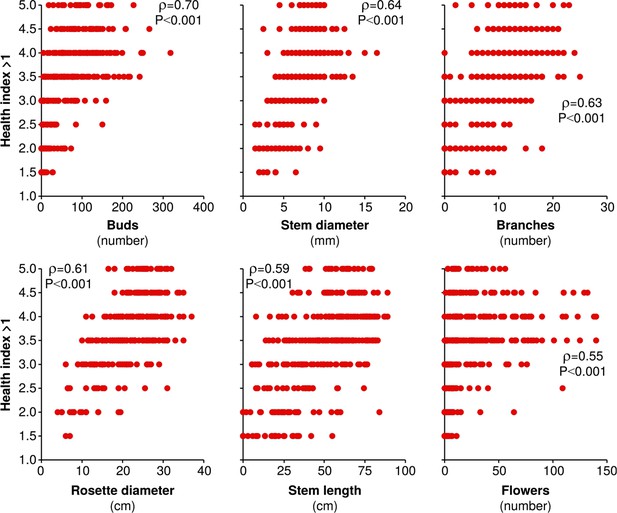
The health index assigned to plants is strongly and significantly correlated to several measures of plant size and reproduction.
All measures of plant size and reproduction made on June 6th in season two are plotted against simultaneously assigned health indices >1 (a health index of 1 refers to dead plants). Spearman's ρ and the corresponding Holm-Bonferroni corrected p-value are shown for each correlation.
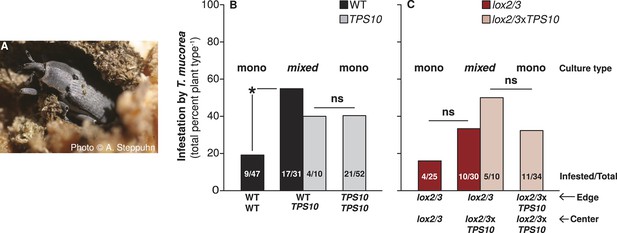
TPS10 plants increase the infestation rates of their WT neighbors with the stem-boring weevil T. mucorea.
(A) Photograph of a T. mucorea adult emerging from the stem which it infested as a larva reprinted with permission from Anke Steppuhn, Copyright 2005. All rights reserved. Adults mate and oviposit on young N. attenuata plants in March and April, and upon hatching, larvae burrow into the growing stem of the plant chosen by their mother (Diezel et al., 2011). Infestation was scored as the number of plants with hollow, frass-filled stems (from which larvae were usually also recovered). (B) T. mucorea infestation more than doubled for WT plants, but did not change for TPS10 plants in mixed cultures vs monocultures. Bars indicate total percentage of plants infested; ratios in bars indicate numbers of infested/total plants. Data were collected at the end of experimental season two (June 29th). *Corrected p<0.05 in a Fisher's exact test, n = 31–47 (all surviving replicates). p-values were corrected for multiple testing using the Holm-Bonferroni method. (C) The presence of lox2/3xTPS10 plants also tended to increase T. mucorea infestation in lox2/3 + lox2/3xTPS10 populations but, likely due to lower replicate numbers caused by higher mortality for plants in lox2/3 and lox2/3xTPS10 monocultures (Figure 9), differences are not significant (corrected p-values=1).
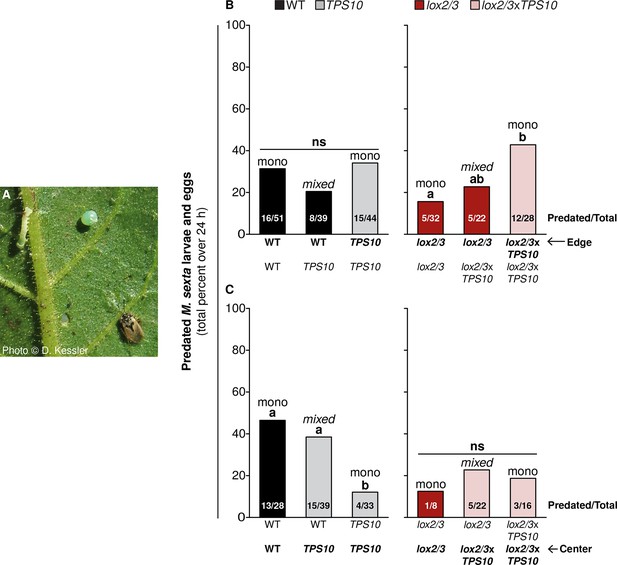
TPS10 and LOX2/3 interact to alter the predation of M. sexta by Geocoris spp from edge vs center plants.
(A) Photograph of a M. sexta egg and first-instar larva, and G. pallens adult on an N. attenuata leaf reprinted with permission from Danny Kessler, Copyright 2006. All rights reserved. As shown here, one first-instar M. sexta larva and one egg were placed on a lower stem leaf in a standardized position on healthy, size-matched plants (one leaf/plant), and their predation was monitored for 24 hr. Data are total numbers from five consecutive trials conducted between June 21st and June 27th at the end of experimental season two, when Geocoris spp. and herbivores were more abundant (see Figure 5). (B and C) Predation rates of M. sexta are higher on lox2/3xTPS10 than lox2/3 plants at the edges of monocultures (B), but lower on TPS10 than WT plants at the centers of monocultures (C). Bars indicate the total percentage of M. sexta eggs and larvae predated per population; numbers of predated/total eggs and larvae are given inside bars. The same trends were observed in both egg and larva predation; separate numbers of larvae and eggs predated are given in Figure 11—source data 1. Plants of the WT background and plants with the lox2/3 silencing construct were separately matched for parallel experiments and thus analyzed separately. (B) Predation of herbivores from edge plants was similar (not significant, ns) regardless of the number of TPS10 plants in WT + TPS10 populations, but increased with increasing numbers of lox2/3xTPS10 plants in lox2/3 + lox2/3xTPS10 populations. For plants at the center of populations, the pattern was reversed: (C) predation rates decreased with increasing numbers of TPS10 plants in WT + TPS10 populations, and were similar (ns) regardless of the number of lox2/3xTPS10 plants in lox2/3 + lox2/3xTPS10 populations. a,b Different letters indicate significant differences (corrected p<0.05) in Fisher's exact tests; p-values were corrected for multiple testing using the Holm-Bonferroni method.
-
Figure 11—source data 1
Total numbers of M. sexta larvae and eggs predated by Geocoris spp. in experimental season two (corresponding to data shown in Figure 10).
- https://doi.org/10.7554/eLife.04490.026
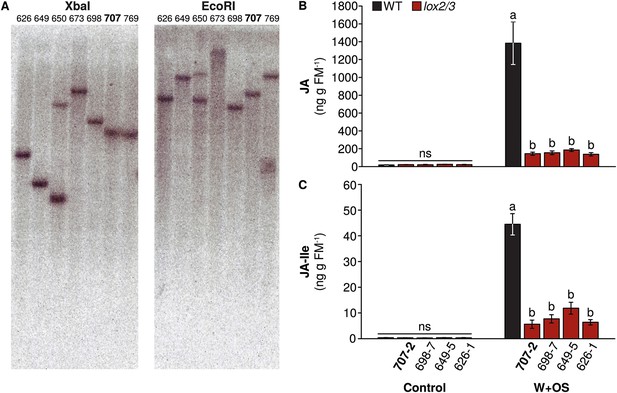
Transgene insertions and reduced jasmonate accumulation in lines of lox2/3.
(A) Southern blot of genomic DNA from multiple lines of lox2/3 digested with the restriction enzymes XbaI or EcoRI. Line 707 is highlighted: this line was used for experiments. A single transgene insertion in TPS10 (line 10–3) is shown in Schuman et al. (2014). All lines of lox2/3 screened had strong reductions in their induced (B) JA and (C) JA-Ile accumulation (mean ± SEM, n = 6). Line 707-2 was used for further experiments. a,b Different letters indicate significant differences (corrected p<0.0001) in Tukey HSD tests following significant (p<0.0001) 1-way ANOVAs on natural log-transformed data (JA: F4,25 = 54.15; JA-Ile: F4,25 = 18.00); ns, not significant in 1-way ANOVAs (p>0.4).
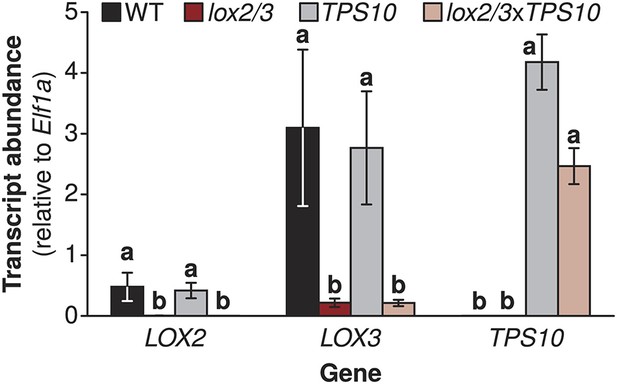
Target gene transcript levels in lox2/3, TPS10, and lox2/3xTPS10 plants.
Transcript accumulation shows that lox2/3 plants, TPS10 plants, and hemizygous crosses (lox2/3xTPS10) have the expected silencing or accumulation of the target genes LOX2, LOX3, and TPS10 (mean ± SEM, n = 4). a,b Different letters indicate significant differences (corrected p<0.01) in Tukey HSD tests following a significant (p<0.001) 1-way ANOVA on natural log-transformed data (LOX2: F3,12 = 49.49, p<0.001; LOX3: F3,12 = 13.73, p<0.001; TPS10: F3,12 = 845.6, p<0.001).
Tables
Volatiles (percent IS plant−1, mean ± SEM) trapped in the headspace around single plants in experimental season one (June 8th–9th).
| GLVs (percent IS plant-1) | TPS10 products (percent IS plant-1) | Non-target volatiles (percent IS plant-1) | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (Z)-Hexen-3-ol | TAB | TBF | α-Duprezianene | Germacrene A | |||||||||||||||||||||||||||||||||
| Genotype | Day n | Night n | Day | Night | Day | Night | Day | Night | Day | Night | Day | Night | |||||||||||||||||||||||||
| WT | 8 | 8 | 0.99% | ± | 0.99% | 7.96% | ± | 4.25% | 0.37% | ± | 0.29% | — | — | — | 2.13% | ± | 0.85% | 0.96% | ± | 0.42% | 1.68% | ± | 0.98% | — | |||||||||||||
| TPS10 | 7 | 7 | 2.37% | ± | 1.55% | 2.84% | ± | 0.63% | 18.97% | ± | 6.03% | 0.94% | ± | 0.53% | 9.34% | ± | 3.44% | 0.20% | ± | 0.20% | 9.47% | ± | 5.26% | 0.94% | ± | 0.26% | 2.78% | ± | 1.52% | — | |||||||
| lox2/3 | 7 | 8 | 0.13% | ± | 0.13% | 1.06% | ± | 0.64% | — | 0.15% | ± | 0.15% | — | — | 2.75% | ± | 1.12% | 0.60% | ± | 0.19% | 1.68% | ± | 1.49% | — | |||||||||||||
| lox2/3xTPS10 | 7 | 7 | 0.07% | ± | 0.07% | 1.24% | ± | 0.84% | 7.39% | ± | 2.56% | 2.08% | ± | 0.84% | 4.47% | ± | 1.70% | 0.40% | ± | 0.40% | 3.02% | ± | 1.42% | 0.73% | ± | 0.31% | 0.66% | ± | 0.37% | — | |||||||
-
, Different letters indicate significant differences between lines for a vertical category (corrected P<0.05) in Wilcoxon rank sum tests following significant Kruskal-Wallis tests across all genotypes (see Results text); P-values were corrected for multiple testing using the Holm-Bonferroni method. Community type was also tested and found not significant. Where there are no letters, there are no significant pairwise differences within a category.
-
, / , Indicate significant differences (corrected P<0.05) between lines with and without either the lox construct (WT and TPS v. lox and loxTPS) or the TPS construct (WT and lox v. TPS and loxTPS) in Wilcoxon rank sum tests for a category; P-values were corrected for multiple testing using the Holm-Bonferroni method. There are no significant pairwise differences in these categories.
-
IS, internal standard; GLVs, green leaf volatiles; TAB, (E)-α-bergamotene; TBF, (E)-β-farnesene; —, not detected.





