Enhanced stability and polyadenylation of select mRNAs support rapid thermogenesis in the brown fat of a hibernator
Peer review process
This article was accepted for publication as part of eLife's original publishing model.
History
- Version of Record published
- Accepted
- Received
Decision letter
-
Benjamin J BlencoweReviewing Editor; University of Toronto, Canada
eLife posts the editorial decision letter and author response on a selection of the published articles (subject to the approval of the authors). An edited version of the letter sent to the authors after peer review is shown, indicating the substantive concerns or comments; minor concerns are not usually shown. Reviewers have the opportunity to discuss the decision before the letter is sent (see review process). Similarly, the author response typically shows only responses to the major concerns raised by the reviewers.
Thank you for sending your work entitled “Enhanced stability and polyadenylation of select mRNAs support rapid thermogenesis in the brown fat of a hibernator” for consideration at eLife. Your article has been evaluated by Randy Schekman (Senior editor) and, a Reviewing editor, and 2 reviewers.
The Reviewing editor and the reviewers discussed their comments before we reached this decision, and the Reviewing editor has assembled the following comments to help you prepare a revised submission.
This manuscript investigates the transcriptome dynamics of hibernating ground squirrels. The authors performed RNA-Seq profiling of brown adipose tissue (BAT) in ground squirrels across different periods of the torpor-arousal cycle. Three major observations were made: (1) unsupervised clustering of the samples revealed three main groups—spring, winter warm, and winter cold. Between these three groups, 1638 genes with significant differential steady-state transcript levels were detected. Gene ontology analysis identified processes associated with BAT function in the winter groups; (2) a subset of transcripts in the winter groups reached their highest abundance during early torpor, late torpor, and early arousal despite suppressed transcription rates at low body temperatures. RT-PCR analysis revealed two classes of transcripts during torpor: transcripts were either degraded and not replenished, or they were stabilized by the maintenance or acquisition of a long poly(A) tail; (3) the authors propose a mathematical model to describe the changes in transcription rates and degradation could affect differential gene expression in torpor. A temperature-dependent mechanism was identified that protected a subset of transcripts relative to bulk RNA degradation.
This study provides novel insight into the transcriptional dynamics of hibernating animals. In particular, how certain transcripts are activated/maintained despite suppressed transcription during low body temperatures is poorly understood. The findings in this study would appeal to those interested in hibernation dynamics, as well as those who study post-transcriptional regulation. While it is known that polyadenylation is important for mRNA stability, localization, and translation, the authors present evidence for the first time that this mechanism is implicated in the torpor-arousal cycle.
Overall, the manuscript reads very well written with clearly reported observations and results. A significant limitation of the manuscript, as was discussed by the three reviewers, is the lack of experimental data addressing the molecular mechanisms underlying the authors' observations.
Main comments:
1) The authors' manuscript would be significantly strengthened if they could provide mechanistic insight into the temperature-dependence of poly(A) tail length. For example, the reviewers wonder whether BAT samples from torpor-arousal cycling animals could be used to assess differential cross-linking/binding of PABP1 and/or RBM3 to transcripts that display polyA length changes, to test the authors' proposal (based on previous observations they have cited) that these factors may be associated with differential transcript stability dynamics. This experiment could potentially be performed using CLIP-Seq, or CLIP-qRT-PCR on selected transcripts, by employing commercial antibodies that bind to conserved epitopes on these proteins (which overall are highly conserved). Related to this, have the authors examined the levels of transcripts from genes that encode factors involved in the control of polyA tail length (i.e. deadenylation or polyadenylation factors) across their RNA-Seq datasets? Such information may lead to mechanistic insight as well.
2) The authors are requested to provide basic information on polyA site usage in the different samples. How many sites were detected and how does the rate of detection of polyA site usage relate to the steady-state expression of the corresponding transcripts?
3) The authors should provide a more basic description (i.e. for the general reader) of how the mathematical model they developed was conceived and how well it reliably models the experimental data. In this regard, it is also not clear in the main text or Methods how levels of bulk and protected transcripts were determined.
https://doi.org/10.7554/eLife.04517.021Author response
1) The authors' manuscript would be significantly strengthened if they could provide mechanistic insight into the temperature-dependence of poly(A) tail length. For example, the reviewers wonder whether BAT samples from torpor-arousal cycling animals could be used to assess differential cross-linking/binding of PABP1 and/or RBM3 to transcripts that display polyA length changes, to test the authors' proposal (based on previous observations they have cited) that these factors may be associated with differential transcript stability dynamics. This experiment could potentially be performed using CLIP-Seq, or CLIP-qRT-PCR on selected transcripts, by employing commercial antibodies that bind to conserved epitopes on these proteins (which overall are highly conserved). Related to this, have the authors examined the levels of transcripts from genes that encode factors involved in the control of polyA tail length (i.e. deadenylation or polyadenylation factors) across their RNA-Seq datasets? Such information may lead to mechanistic insight as well.
We agree that our manuscript could be strengthened by providing mechanistic insight into the temperature-dependence of poly(A) tail length and selective transcript stability. Although the suggested strategy of either CLIP or CLIP-Seq using antibodies to PABP1 or RBM3 might enhance our understanding of the molecular mechanism involved, this is often not a trivial undertaking in ground squirrels in terms of identifying useful antibodies. We do not think that evidence for PABP1 binding would be particularly helpful because it is unlikely to provide the specificity needed to explain the selectivity among transcripts that is evident in our data. Before embarking on a potentially difficult search for an appropriate immunoprecipitating RBM3 antibody, we employed a motif-searching bioinformatics approach to see whether RBM3 binding sites were enriched in the protected transcript subset. Although neither of the two (worrisomely unrelated) RBM3 binding sites that have been reported by others are significantly enriched in our population of protected transcripts (see Author response image 1), we believe that the motif searching strategy employed set us on a path that has proven to be extremely fruitful. Specifically, we detected two C-rich motifs that are significantly enriched in the 3’ regions of the stabilized transcripts; these motifs were the only enriched motifs (see figure) and are predicted to be bound by a poly(C) binding protein (PCBP). Significantly, the mRNA expression for one of the PCBP family members, PCBP3, increased during the winter in our dataset, in contrast to most of the other RNA binding proteins that were differentially expressed. We document this motif and its link with PCBP3 in a new Figure in the manuscript (Figure 6) and now suggest, based on the motif enrichment analysis, that a PCBP paralog, most likely PCBP3, may be mechanistically linked to the observed transcript stability. We believe this analysis adds significant mechanistic insight underlying mRNA stabilization and polyadenylation and paves the way for the next step, which would be to identify the RNAs that are bound by PCBP3 (or its paralogs) at different hibernation timepoints using CLIP-Seq. We would like to argue here, however, that such a next step is well beyond the scope of this study.
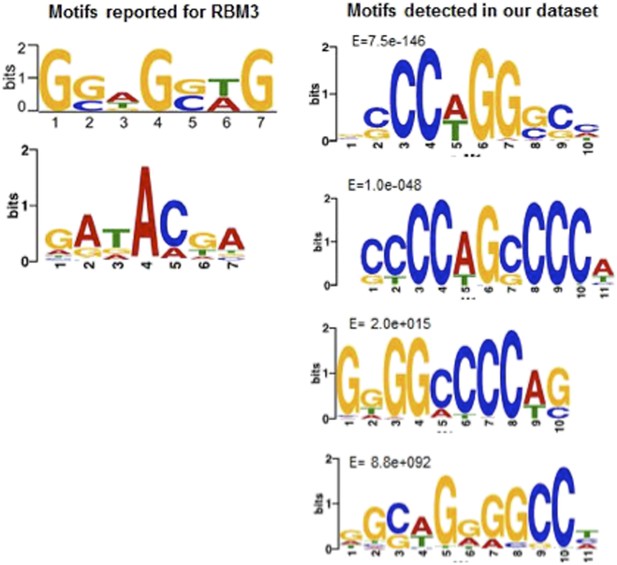
Motif comparisons: Left side, RBM3 motifs reported by (top) Liu, Y. et al. (2013) Sci. Rep. 3, doi:10.1038; (bottom); Ray, D. et al. (2013), Nature 499, 172. Right side, top four motifs in the hibernation-increased subset of RNAs in our dataset. Note only the top two, as reported in the new Figure 6 of our manuscript, are significantly enriched.
2) The authors are requested to provide basic information on polyA site usage in the different samples. How many sites were detected and how does the rate of detection of polyA site usage relate to the steady-state expression of the corresponding transcripts?
We hesitate to make any inference regarding poly(A) site usage at this point. If NlaIII cut to completion, we at best have identified the 3’ most “CATG” site within each transcript, not the 3’ end itself. Thus, we do not think these tags should be viewed as a conclusive proxy for defining 3’UTR’s and poly(A) site usage. However, we do provide all tag locations within their respective transcripts in the supplementary tables, so that others doing poly(A)-Seq analysis on ground-squirrel transcripts can easily make such comparisons. Our observations of enhanced mRNA stabilization and polyadenylation during torpor were an unanticipated finding from the digital transcriptome data that were collected to identify differentially-expressed genes linked to the phenotypic dynamics of hibernation. Although in the process we have detected what we suspect are alternative polyadenylation sites, focusing on them here would detract from the main findings of this work.
3) The authors should provide a more basic description (i.e. for the general reader) of how the mathematical model they developed was conceived and how well it reliably models the experimental data. In this regard, it is also not clear in the main text or Methods how levels of bulk and protected transcripts were determined.
We made several changes throughout the manuscript to enhance this information in the main text.
https://doi.org/10.7554/eLife.04517.022




