Early asymmetric cues triggering the dorsal/ventral gene regulatory network of the sea urchin embryo
Figures
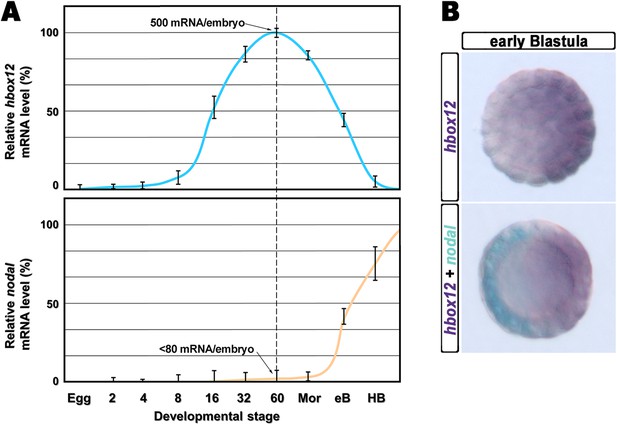
Expression of hbox12 and nodal genes during early embryogenesis of P. lividus.
(A) Temporal expression profiles examined by qPCR. Values at the different stages are shown as a percentage of the maximum signal intensity. Absolute numbers of transcripts per embryo given at the 60-cell stage are averages of the results of two independent experiments using distinct batches of cDNA. Abbreviations of the examined developmental stages: 2, 2-cell; 4, 4-cell; 8, 8-cell; 16, 16-cell; 32, 32-cell; 60, 60-cell; Mor, morula; eB, early blastula; HB, hatching blastula. (B) Spatial restriction of the hbox12 and nodal transcripts observed following WMISH at the indicated stage.
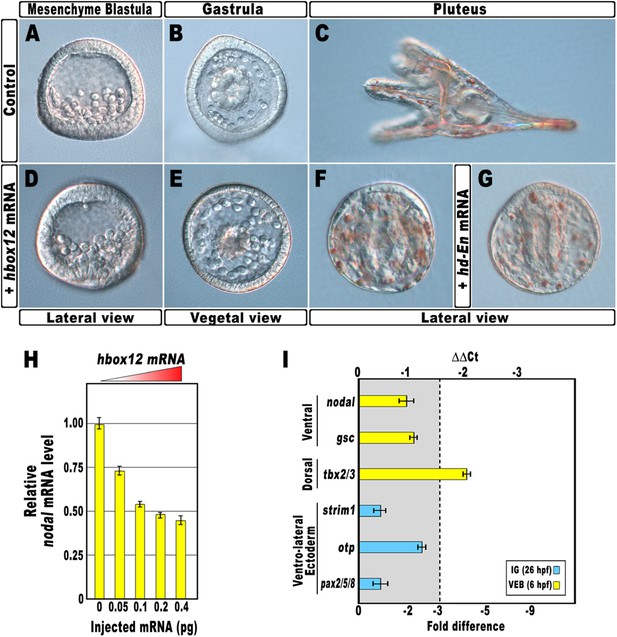
Disruption of embryonic DV polarity by ectopic expression of hbox12.
(A–G) 0.1 pg of the full-length hbox12 mRNA (D–F) or the hd-En mRNA (G), as well as a control out-of-frame strim1 transcript (A–C), were injected into zygotes and embryos were observed at the indicated stages. Overexpression of either hbox12 or hd-En severely perturbed DV axis formation, inflicting morphological defects that appeared from the gastrula stage onward. (H) qPCR measurements of nodal transcript abundance in embryos injected with increasing amounts of the hbox12 mRNA. Values are shown as a percentage of the nodal mRNA level in control uninjected embryos. Further detail for the qPCR procedure is given in ‘Materials and methods’. (I) Changes in gene expression level of nodal and other territorial marker genes assessed by qPCR in hbox12-injected embryos. Data are indicated as normalized ΔCt (ΔΔCt, left ordinate), and as the corresponding fold difference in transcript abundance (right ordinate), with respect to control embryos, at the same stage of development, derived from zygotes injected with the strim1 out-of-frame transcript. The gray region represents ΔΔCt values corresponding to less than threefold difference. Although this is commonly considered the limit of significance for qPCR assays, the relevance of our measurements is reinforced by WMISH results (see text for details). Error bars are standard errors for the qPCR replicates. Oligonucleotide primer pairs used for qPCR reactions and amplicon lengths are indicated in Supplementary file 1. Abbreviations: lG, late gastrula; VEB, very early blastula.
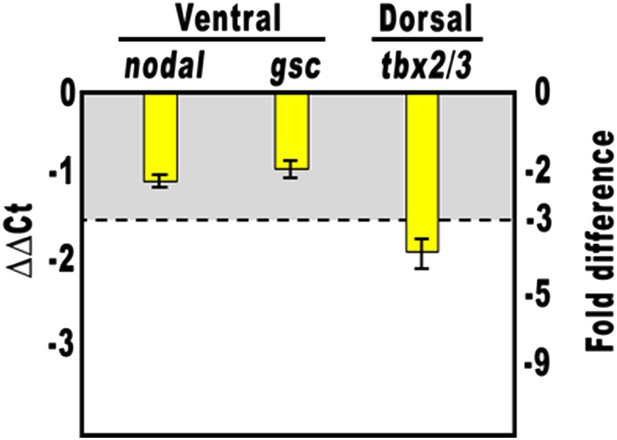
Overexpression of the HD-En obligate repressor and effect on nodal and gsc gene expression.
Changes in gene expression levels was assessed by qPCR in cDNA samples resulting from hd-En-injected embryos. Data are normalized and indicated as in Figure 2I. The gray region represents ΔΔCt values corresponding to less than threefold difference. See also Supplementary file 1.
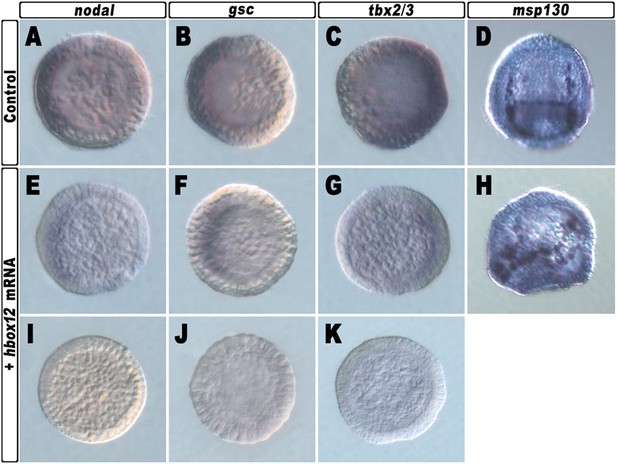
Spatial distribution of ectoderm- and PMC-specific markers in control and hbox12 overexpressing embryos.
Control strim1 out-of-frame transcript (A–D) and hbox12 mRNA injected (E–K) embryos were fixed at the mesenchyme blastula (A–C, E–G and I–K) or prism (D and H) stages and analysed by chromogenic WMISH with the indicated probes. The embryos shown in (D) and (H) are oriented in a oral/ventral and lateral view, respectively, while all the other embryos are in a vegetal view.
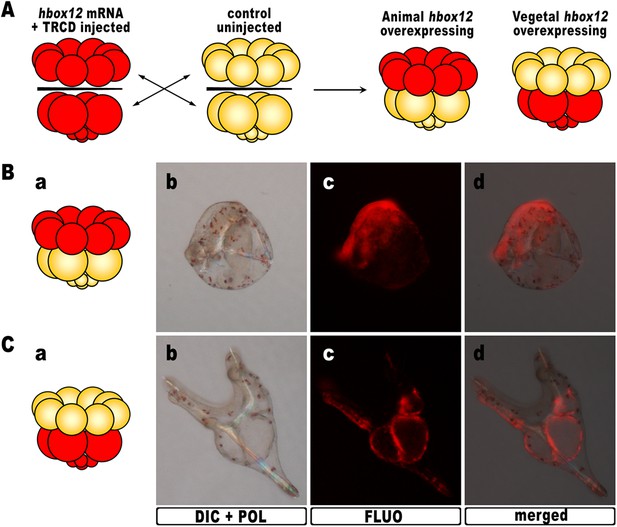
Overexpression of hbox12 in chimeric embryos.
(A) Schematic illustrating that, at the 16-cell stage, animal and vegetal halves of hbox12/TRCD-injected and control uninjected embryos were isolated and recombined. (B–C) Side views of representative examples of the resulting reciprocal chimeras examined at 48 hpf. The composition of the chimeras is shown in the diagrams in the left panels (Ba and Ca). Images for each embryo are shown under DIC optics with simultaneous plane polarized light illumination (Bb and Cb), epifluorescence (Bc and Cc), and aggregate merging (Bd and Cd).
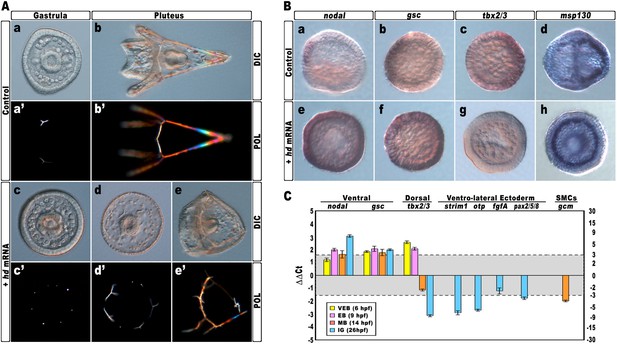
Impairing hbox12 function and effects on DV axis formation.
(A) Zygotes were injected with the control out-of-frame strim1 RNA (Aa–Ab) or the hd mRNA (Ac–Ae), and the resulting embryos were observed from a vegetal view at the indicated stages. (B) Control- (Ba–Bd) and hd-injected (Be–Bh) embryos were fixed at the mesenchyme blastula stage and analysed by WMISH with the indicated probes. The embryo shown in (Ba) is oriented in a lateral view, while all the other embryos are in a vegetal view. (C) Changes in gene expression level of territorial marker genes assessed by qPCR during development of hd-injected embryos. Data are normalized and indicated as in Figure 2I. The gray region represents ΔΔCt values corresponding to non-significant variation (less than threefold difference). See also Supplementary file 1. Abbreviations: VEB, very early blastula; EB, early blastula; MB, mesenchyme blastula; lG, late gastrula.
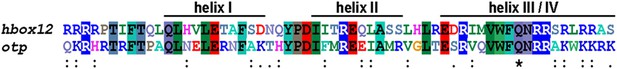
Multiple comparison of the homeodomain of Hbox12 and Otp proteins from P. lividus.
Identical residues are shaded by differently coloured boxes, while double and single dots indicate decreasing degree of conservation; divergent amino acids are indicated by blank spaces. The Glutamine at position 50 is marked by an asterisk. Accession numbers were: Hbox12, X83675.1, and Otp, AJ007501.1.
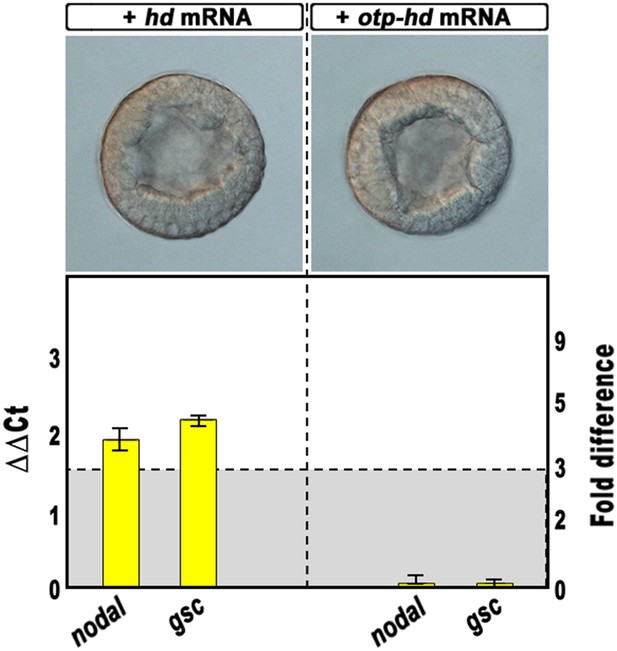
Overexpression of isolated homeodomains and effect on nodal and gsc gene transcription.
Zygotes were injected with the synthetic mRNAs coding for the isolated homeodomain of either Hbox12 or Otp and observed at the early blastula stage (upper panels). Total RNA was extracted from embryos at the morula stage, and changes in gene expression level of nodal and gsc determined by qPCR measurements. Data are indicated as in Figure 2I. The gray region represents ΔΔCt values corresponding to non-significant variation (less than threefold difference). See also Supplementary file 1.
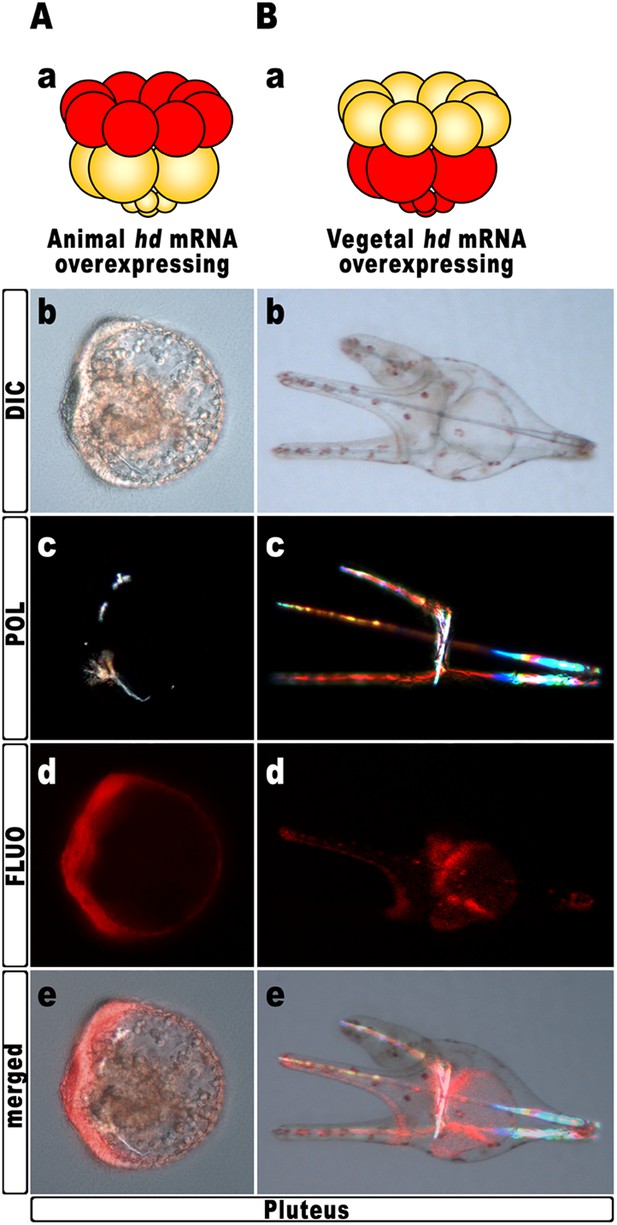
Block of hbox12 function in the animal hemisphere of chimeric embryos, and effects on DV patterning.
(A–B) Diagrams in (Aa) and (Ba) show the composition of the reciprocal chimeras resulting from animal and vegetal half recombination of hd/TRCD-injected and control uninjected embryos at the 16-cell stage. (Ab–Ae and Bb–Be) Side views of the resulting chimeras examined at 48 hpf. Images for each embryo are shown under DIC optics (Ab and Bb), plane polarized light illumination (Ac and Bc), epifluorescence (Ad and Bd), and aggregate merging (Ae and Be).
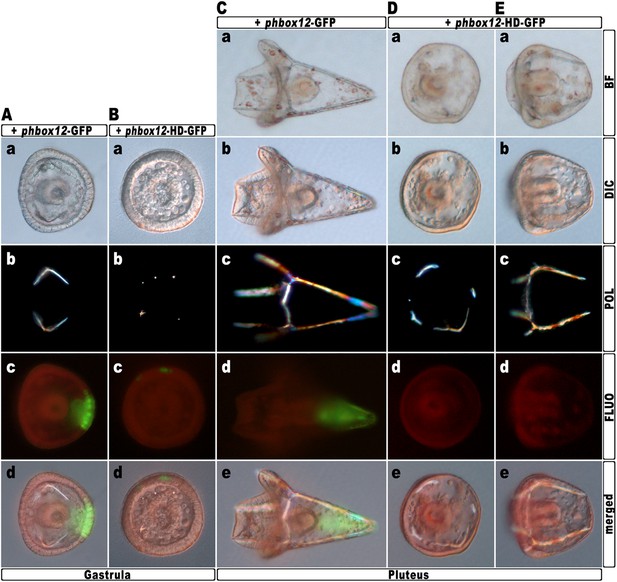
Block of hbox12 function in aboral ectoderm cells of transgenic embryos, and effects on DV patterning.
(A–E) Zygotes were injected with the indicated transgenes, and the resulting embryos were observed from a vegetal view at the indicated stages. Bright field (Ca, Da, and Ea), DIC (Aa, Ba, Cb, Db, and Eb), Dark field (Ab, Bb, Cc, Dc, and Ec), epifluorescence (Ac, Bc, Cd, Dd, and Ed), and aggregate merging (Ad, Bd, Ce, De, and Ee) images are shown.
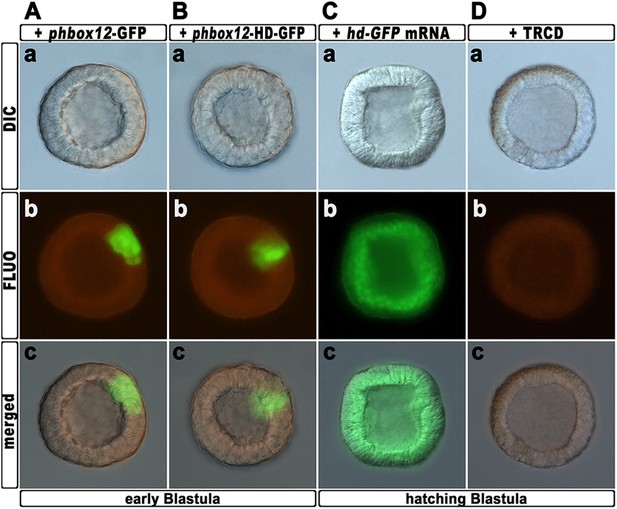
Expression of the HD-GFP fusion protein during early embryogenesis.
(A–C) Zygotes were injected with either the phbox12-GFP (A) or phbox12-HD-GFP (B) DNA constructs or with the hd-GFP synthetic transcript (C), and embryos were observed at the indicated stages. (D) Control embryo injected with TRCD.
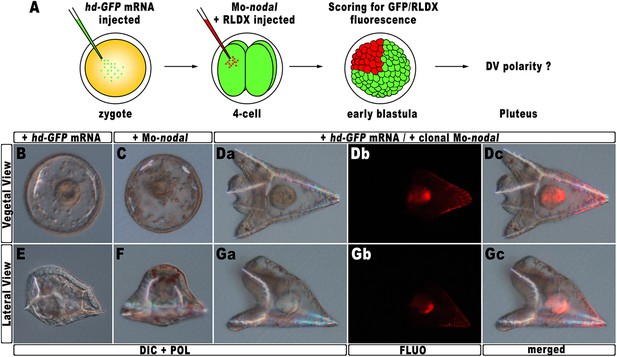
Rescue of DV polarity by clonal knock-down of nodal into hd-injected embryos.
(A) At the 4-cell stage, one blastomere of hd-GFP mRNA injected embryos was co-injected with a morpholino oligonucleotide directed against nodal (Mo-nodal) together with the RLDX red fluorescent tracer. The resulting embryos were scored for simultaneous GFP/RLDX fluorescence at the early blastula stage, and eventually examined by microscopic observation at the pluteus stage. (B–G) Representative examples of embryos injected with either the hd-GFP mRNA (B and E) or the Mo-nodal alone (C and F), and of double-injected rescued embryos (D–G). Note that in both the rescued pluteus larvae shown in (D) and (G), the progeny of the blastomere that received Mo-nodal was embedded into the dorsal structures.
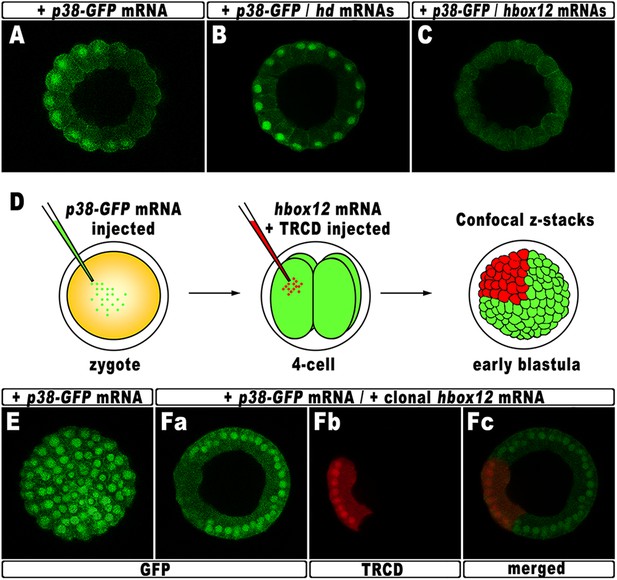
Functional correlation between hbox12 and p38 MAPK.
(A–C) Embryos injected with the p38-GFP mRNA either alone (A) or in the presence of the hd mRNA (B) or the hbox12 mRNA (C), respectively, and analyzed by live confocal microscopy at the 60-cell stage. Images are internal projections of 10–15 sections from a confocal z-series. (D) At the 4-cell stage, one blastomere of p38-GFP mRNA injected embryos was co-injected with the hbox12 mRNA, and the resulting embryos were examined by live confocal microscopy at the early blastula stage. Representative examples of control p38-GFP mRNA injected (E) and double-injected (F) embryos are shown.
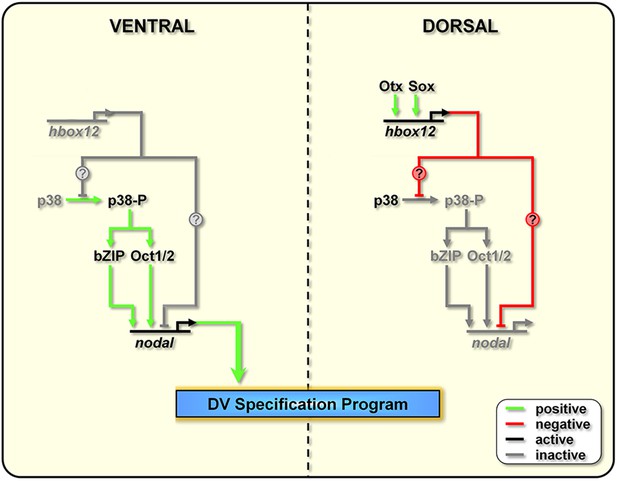
Model for establishment of the DV organizing centre in the sea urchin embryo.
In the early embryo, hbox12 transcription is initiated by combinatorial positive inputs from Otx and probably Sox in the future dorsal ectoderm (Cavalieri et al., 2008). hbox12-dependent suppression of nodal gene expression in these cells is mediated by the transient inactivation of p38 and/or probably by direct repression. On the ventral side of the embryo, hbox12 expression is negatively regulated by unidentified repressors (Cavalieri et al., 2008). In these cells, active p38 stimulates nodal expression probably through Oct1/2 or other intermediate transcription factors (Range and Lepage, 2011), allowing the establishment of the DV organizer and patterning along the secondary axis.
Additional files
-
Supplementary file 1
List of gene specific oligonucleotides used in the quantitative RT-PCR.
- https://doi.org/10.7554/eLife.04664.017





