Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery
Figures
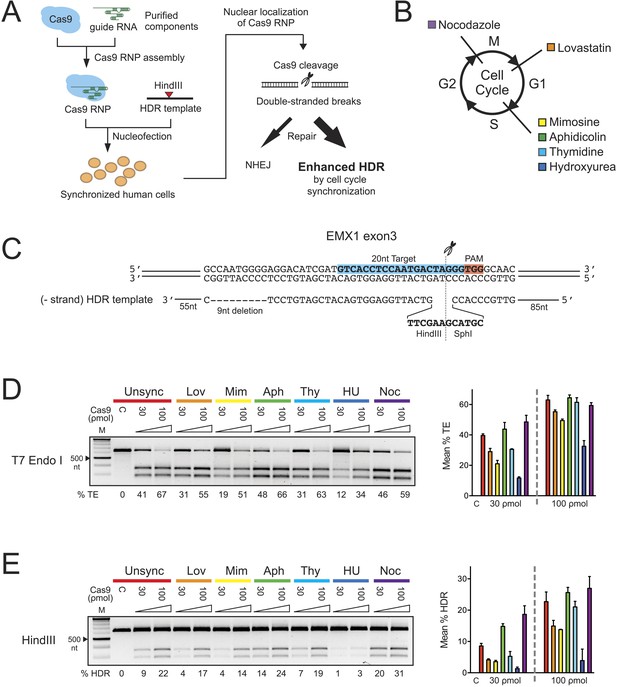
The effect of cell cycle synchronization on total editing and homology-directed repair frequencies in HEK293T cells.
(A) Experimental schematic of timed delivery of Cas9-guide RNA ribonucleoprotein (RNP) into human cells for genome editing. (B) Chemical inhibitors used to arrest cells at specific phases of cell cycle included lovastatin (Lov), which blocks at early G1 and partially at G2/M phase; mimosine (Mim), aphidicolin (Aph), thymidine (Thy) and hydroxyurea (HU) which arrest cells at the G1-S border prior to onset of DNA replication; and nocodazole (Noc) which causes arrest at G2/M phase. (C) The homology-directed repair (HDR) donor DNA is a 183 nt ssODNA that is complementary to the target sequence (−strand) and contains a 9 nt insertion (HindIII and SphI restriction sequences) at the cut site and a 9 nt deletion downstream of the cut site; these modifications are flanked by 85 nt and 55 nt asymmetrical homology arms at 5′ and 3′ ends, respectively. (D, E) PCR-based screening of cell cycle inhibitors for enhancement of Cas9-triggered total editing (TE) (D) and HDR (E) frequencies in HEK293T cells. For each inhibitor condition (color coded), two doses of Cas9 RNP, 30 and 100 ρmol, were transfected with 100 ρmol of HDR DNA template; control reactions (labeled as C) contained 100 ρmol of Cas9 but no sgRNA. The TE frequency was measured using a T7 endonuclease I assay and analyzed using a formula described in ‘Materials and Methods’. The HDR frequency was determined directly by HindIII digestion, which specifically cleaved the newly integrated HindIII sequence, and calculated as the ratio of DNA product to DNA substrate. The % TE, % HDR and standard deviation (error bars) were calculated from three experiments.
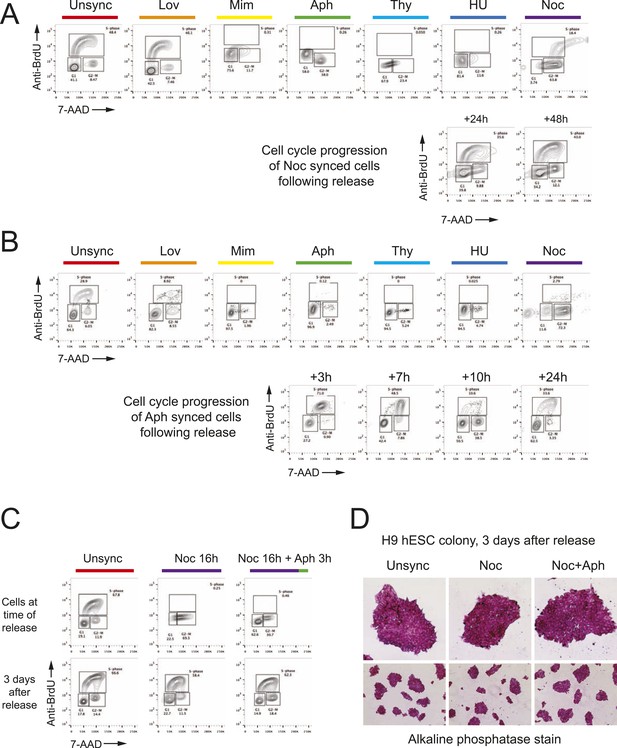
FACS analysis reveals cell cycle blocks and the DNA content in the cells that are arrested at different phases of cell cycle.
Bivariate cell cycle, BrdU (S-phase), 7-AAD (DNA content) FACS analysis reveals cells are arrested at different phases of cell cycle. Chemical inhibitors were used to arrest cells at specific phases of cell cycle. (A) Analysis of HEK293T cells with different cell cycle blocks and nocodazole released cells. (B) Analysis of human neonatal fibroblasts with different cell cycle blocks and aphidicolin released cells. (C) Analysis of H9 hES cells unsynchronized (unsync), nocodazole synchronzied (Noc) and nocodazole + aphidicolin sequential synchronized (Noc + Aph) at time of cell cycle block and transfection or 3 days after release. Alkaline phosphatase positive and normal ES colony morphology for all three conditions. ROCK apoptosis inhibitor (10 μM) was required for survival of synchronized H9 ESCs after release when cultured at low density in 6-well plates but not when cultured at high density in 96-well plates.
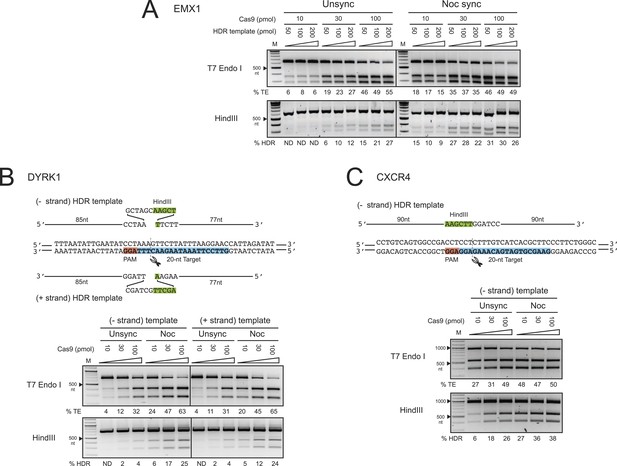
The enhancement of TE and HDR at the EMX1, DYRK1 and CXCR4 loci by nocodazole synchronization in HEK293T cells.
(A) The effect of nocodazole on the TE and HDR frequencies at EMX1 locus. HEK293T cells were synchronized at M phase with 200 ng/ml of nocodazole for 17 hr before nucleofection. To determine the optimal dosage, three concentrations of Cas9 RNP were assayed in combination with three doses of HDR template (Figure 1C). The TE frequencies at 10 ρmol of Cas9 RNP in the unsynchronized cells were too low and therefore not determined (ND). (B) The effect of nocodazole on the TE and HDR frequencies at DYRK1 locus. The directionality of ssODNA HDR templates, either identical (+strand) or complementary (−strand) to the target sequence, was examined. The PAM is highlighted in red, the target sequence in blue and the integrated HindIII site in green. (C) The effect of nocodazole on the TE and HDR frequencies at the CXCR4 locus. The HDR template is ssODNA complementary (−strand) to the target sequence, and contains a HindIII restriction sequence flanked by 90 nt homology arms. Representative gels from two biological replicates are shown.
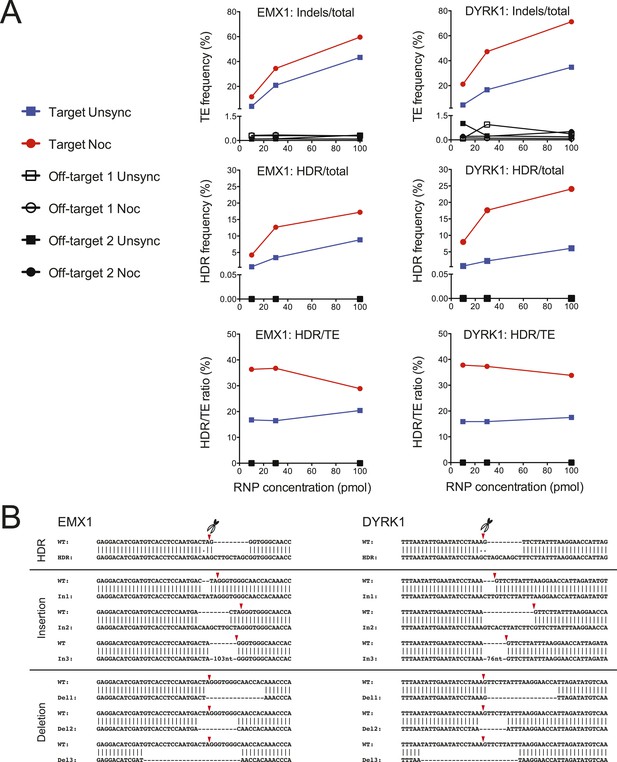
On-target NHEJ and HDR and off-target cleavage analyses by deep sequencing.
(A) The genomic DNAs from Figure 2A,B experiments were analyzed for NHEJ and HDR frequencies, at the on-target and off-target sites, by deep sequencing. The TE frequency (indels/total reads) was determined at the EMX1 target, DYRK1 target and selected off-target loci as a function of Cas9 RNP dosage, (n = 1 representative experiment). The HDR frequency (HDR/total reads) represented specifically exogenous donor template-mediate HDR. The ratio of Cas9 RNP-induced DSB repaired by the HDR pathway was determined as the percentage of HDR/TE. The controls, which included the non-transfected cells and the cells transfected with only Cas9 protein but no sgRNA, showed no evidence of on- or off-target editing. (B) Representative sequences repaired by HDR and NHEJ at the EMX1 and DYRK1 loci. The Cas9 cleavage sites are marked by red triangles.
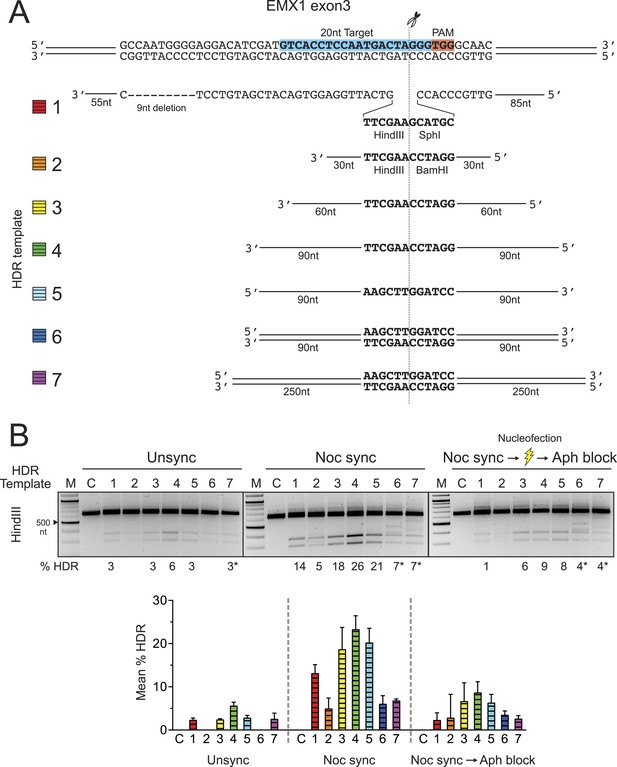
Systematic investigation of DNA templates for efficient HDR at the EMX1 locus in HEK293T cells.
(A) Segment of human EMX1 exon 3 shows the 20 nt target sequence (highlighted in blue), the TGG PAM region (in red) and the Cas9 cleavage site at three bases upstream from PAM. Seven HDR templates (color coded) were tested for HDR efficiency. Template 1 is as described in Figure 1C. Templates 2–7 contain HindIII and BamHI restriction sites that are flanked symmetrically by various lengths of homology arms, ranging from 30 nt to 250 nt. Templates 2–5 are ssODNA; templates 6–7 are PCR amplified double-stranded DNA (see ‘Materials and methods’). (B) HDR efficiency was tested under three cell conditions. In addition the unsynchronized and nocodazole synchronized conditions, the cells were synchronized with nocodazole prior to nucleofection, and immediately post nucleofection, a single dose of aphidicolin (2 μg/ml) was added to the growth media to prevent the transfected cells from proceeding into the S phase. The purpose was to test whether blocking passage through S phase reduces HDR efficiency, since the HDR pathway is thought to be most active during S phase. This one-time addition of aphidicolin is labeled as ‘Aph block’ in the third panel, as opposed to the standard aphidicolin synchronization procedure used elsewhere in the manuscript. Thirty ρmol of Cas9 RNP and 50 ρmol of HDR template were used in the nucleofection reaction; the control reaction (C) contained no HDR template. The mean % HDR and standard deviation (error bar) was determined by HindIII digestion from three experiments. Representative gels from PCR and HDR analyses are shown for each cell condition. Templates 6 and 7 produced unusual banding patterns, making quantitation of DNA bands less accurate (labeled by asterisk).
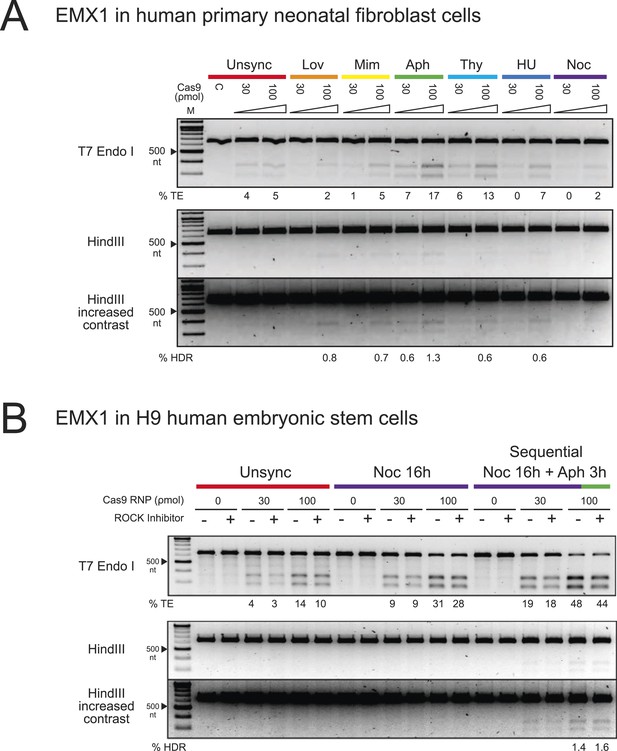
The enhancement of TE and HDR frequencies at the EMX1 locus by cell cycle synchronization in human primary neonatal fibroblast and embryonic stem cells.
(A) Screening of cell cycle inhibitors for enhancement of TE and HDR frequencies in human primary neonatal fibroblast cells. For each inhibitor condition (color coded), two doses of Cas9 RNP, 30 and 100 ρmol, were transfected with 100 ρmol of HDR DNA template 4 from Figure 3A. A control reaction (labeled as C) contained 100 ρmol of Cas9 but no sgRNA. The % TE and % HDR were analyzed similarly as with HEK293T cells. (B) Three conditions were tested using hES cells: unsynchronized, nocodazole synchronized and nocodazole-aphidicolin sequential synchronized. The cells were treated with nocodazole for 16 hr, washed to remove the drug and then treated with aphidicolin for 3 hr before nucleofection. 30 or 100 ρmol of Cas9 RNP was co-transfected with 100 ρmol of HDR template 4 from Figure 3A, and cultured at high density in 96-well plates in the presence or absence of ROCK apoptosis inhibitor (10 μM). For both experiments, representative gels from two biological replicates are shown. The contrast of the gel images was increased to show that no HDR was detected in other conditions.
Additional files
-
Supplementary file 1
PCR primers for target loci amplification and indexing for deep sequencing of on-target and off-target sites. Also included is deep sequencing data and analysis.
- https://doi.org/10.7554/eLife.04766.008





